8IX3
 
 | | Cryo-EM structure of SARS-CoV-2 BA.4/5 spike protein in complex with 1G11 (local refinement) | | Descriptor: | BA.4/5 variant spike protein, heavy chain of 1G11, light chain of 1G11 | | Authors: | Sun, H, Jiang, Y, Zheng, Z, Zheng, Q, Li, S. | | Deposit date: | 2023-03-31 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural basis for broad neutralization of human antibody against Omicron sublineages and evasion by XBB variant.
J.Virol., 97, 2023
|
|
8U1T
 
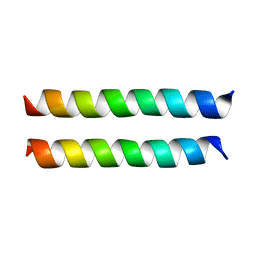 | | SARS-CoV-2 Envelope Protein Transmembrane Domain: Dimeric Structure Determined by Solid-State NMR | | Descriptor: | Envelope small membrane protein | | Authors: | Zhang, R, Qin, H, Prasad, R, Fu, R, Zhou, H.X, Cross, T. | | Deposit date: | 2023-09-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Dimeric Transmembrane Structure of the SARS-CoV-2 E Protein.
Commun Biol, 6, 2023
|
|
8U28
 
 | |
8FIG
 
 | |
8SQ9
 
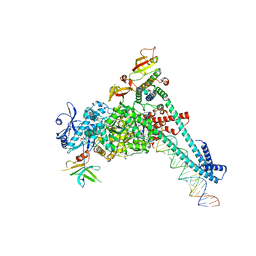 | | SARS-CoV-2 replication-transcription complex bound to nsp9 and UMPCPP, as a pre-catalytic NMPylation intermediate | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]uridine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8SQJ
 
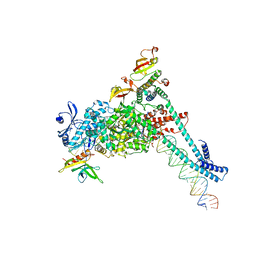 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9, as a noncatalytic RNA-nsp9 binding mode | | Descriptor: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8SQK
 
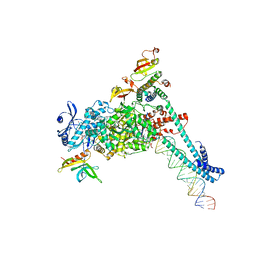 | | SARS-CoV-2 replication-transcription complex bound to RNA-nsp9 and GDP-betaS, as a pre-catalytic deRNAylation/mRNA capping intermediate | | Descriptor: | 5'-O-[(R)-hydroxy(thiophosphonooxy)phosphoryl]guanosine, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Small, G.I, Darst, S.A, Campbell, E.A. | | Deposit date: | 2023-05-04 | | Release date: | 2023-11-22 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural and functional insights into the enzymatic plasticity of the SARS-CoV-2 NiRAN domain.
Mol.Cell, 83, 2023
|
|
8X1H
 
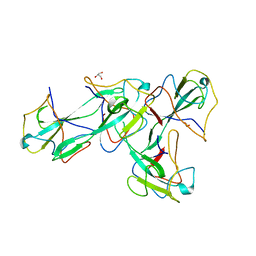 | |
7YBN
 
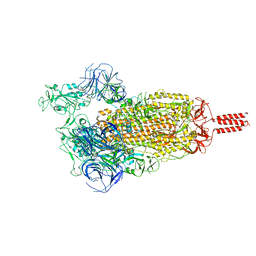 | | SARS-CoV-2 C.1.2 variant spike (Open state) | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
8I4S
 
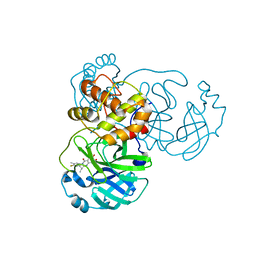 | | the complex structure of SARS-CoV-2 Mpro with D8 | | Descriptor: | 3-(4-fluoranyl-3-methyl-phenyl)-2-(2-methylpropyl)-5,6,7-tris(oxidanyl)quinazolin-4-one, ORF1a polyprotein | | Authors: | Lu, M. | | Deposit date: | 2023-01-21 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of quinazolin-4-one-based non-covalent inhibitors targeting the severe acute respiratory syndrome coronavirus 2 main protease (SARS-CoV-2 M pro ).
Eur.J.Med.Chem., 257, 2023
|
|
8J1T
 
 | |
8J1V
 
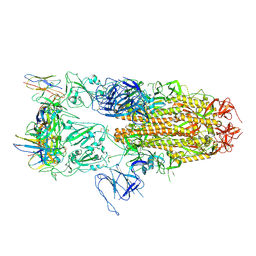 | |
8R0V
 
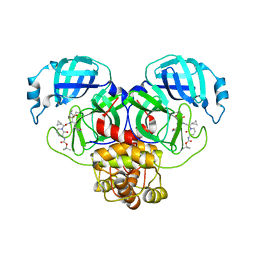 | | SARS-CoV-2 Mpro (Omicron, P132H) in complex with alpha-ketoamide 13b-K at pH 6.5 | | Descriptor: | 3C-like proteinase nsp5, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclopropyl-1-oxidanylidene-1-[[(2~{S},3~{R})-3-oxidanyl-4-oxidanylidene-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]-4-[(phenylmethyl)amino]butan-2-yl]amino]propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Ibrahim, M, Sun, X, Hilgenfeld, R. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|
8R19
 
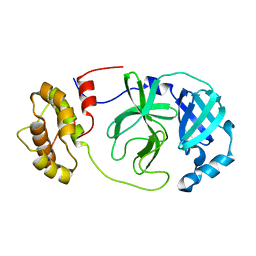 | | SARS-CoV-2 Mpro (Omicron, P132H) free enzyme | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Ibrahim, M, Sun, X, Hilgenfeld, R. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Why is the Omicron main protease of SARS-CoV-2 less stable than its wild-type counterpart? A crystallographic, biophysical, and theoretical study
Hlife, 2, 2024
|
|
8T5P
 
 | |
8T5Q
 
 | |
8UUL
 
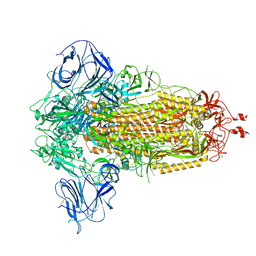 | | Prototypic SARS-CoV-2 spike (containing K417) in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UUM
 
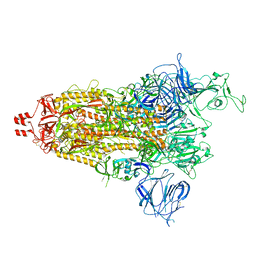 | | Prototypic SARS-CoV-2 spike (containing K417) in the open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose, ... | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UUN
 
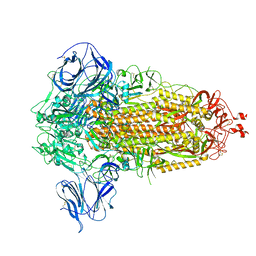 | | Prototypic SARS-CoV-2 spike (containing V417) in the closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8UUO
 
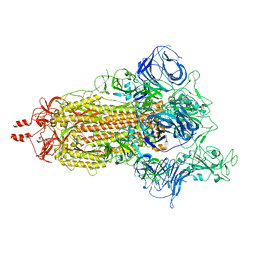 | | Prototypic SARS-CoV-2 spike (containing V417) in the open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, alpha-D-mannopyranose, ... | | Authors: | Geng, Q, Liu, B, Li, F. | | Deposit date: | 2023-11-01 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Lys417 acts as a molecular switch that regulates the conformation of SARS-CoV-2 spike protein.
Elife, 12, 2023
|
|
8WRH
 
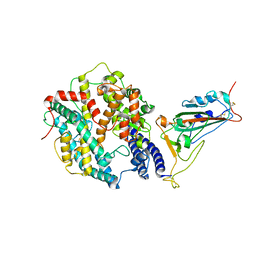 | | SARS-CoV-2 XBB.1.5.70 in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S2' | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-14 | | Release date: | 2023-11-29 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
8WRL
 
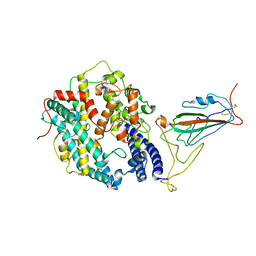 | | XBB.1.5 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Feng, L.L, Feng, L.L. | | Deposit date: | 2023-10-15 | | Release date: | 2023-11-29 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding.
Plos Pathog., 19, 2023
|
|
7YC2
 
 | |
8FTC
 
 | | Crystal structure of main protease of SARS-CoV-2 complexed with inhibitor | | Descriptor: | (1R,2S,5S)-3-[N-(difluoroacetyl)-3-methyl-L-valyl]-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopiperidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Chen, P, Khan, M.B, Lu, J, Arutyunova, E, Young, H.S, Lemieux, M.J. | | Deposit date: | 2023-01-11 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Effect of Deuteration and Homologation of the Lactam Ring of Nirmatrelvir on Its Biochemical Properties and Oxidative Metabolism.
Acs Bio Med Chem Au, 3, 2023
|
|
8HWS
 
 | | The complex structure of Omicron BA.4 RBD with BD604, S309, and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab Heavy chain, BD-604 Fab Light chain, ... | | Authors: | He, Q.W, Xu, Z.P, Xie, Y.F. | | Deposit date: | 2023-01-02 | | Release date: | 2023-12-06 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | An updated atlas of antibody evasion by SARS-CoV-2 Omicron sub-variants including BQ.1.1 and XBB.
Cell Rep Med, 4, 2023
|
|








