9RXL
 
 | | SARS-CoV-2 nucleocapsid C-terminal domain in complex with BCY00018176 | | Descriptor: | 1-[3,5-bis(3-bromanylpropanoyl)-1,3,5-triazinan-1-yl]-3-bromanyl-propan-1-one, BCY00018176, GLYCEROL, ... | | Authors: | Brear, P, Lulla, A, Dods, R, Bezerra, G.A, Hyvonen, M. | | Deposit date: | 2025-07-11 | | Release date: | 2026-03-04 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Utilizing Constrained Bicyclic Peptides for In Vitro Diagnostics.
Acs Nano, 2026
|
|
9KH3
 
 | | Crystal structure of SARS-Cov-2 main protease Y54C mutant in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2024-11-09 | | Release date: | 2026-03-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
Y54C mutant in complex with S217622
To Be Published
|
|
9KH1
 
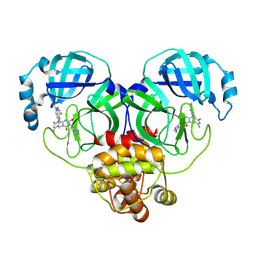 | | Crystal structure of SARS-Cov-2 main protease V186F mutant in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2024-11-09 | | Release date: | 2026-03-04 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
V186F mutant in complex with S217622
To Be Published
|
|
28WF
 
 | |
9KH0
 
 | | Crystal structure of SARS-Cov-2 main protease S46F mutant in complex with S217622 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zhou, X.L, Zhang, J, Li, J. | | Deposit date: | 2024-11-09 | | Release date: | 2026-03-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
S46F mutant in complex with S217622
To Be Published
|
|
9P6F
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor SR-B-78 | | Descriptor: | (1R,2S,5S)-N-[(1S,2Z)-2-imino-1-(5-methoxypyridin-3-yl)ethyl]-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Blankenship, L.R, Liu, W.R. | | Deposit date: | 2025-06-18 | | Release date: | 2026-03-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | From nicotine to SARS-CoV-2 antivirals with potent in vivo efficacy and a broad anti-coronavirus spectrum.
Nat Commun, 2026
|
|
9OBH
 
 | | Co-Structure of SARS-CoV-2 with Compound 34 | | Descriptor: | (4S)-4-(iminomethyl)-3-(isoquinolin-4-yl)-1-[(1s,3R)-3-(trifluoromethyl)cyclobutyl]imidazolidin-2-one, 3C-like proteinase nsp5 | | Authors: | Knapp, M.S, Ornelas, E. | | Deposit date: | 2025-04-22 | | Release date: | 2026-03-04 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | Discovery of EGT710, an Oral Nonpeptidomimetic Reversible Covalent SARS-CoV-2 Main Protease Inhibitor.
J.Med.Chem., 2026
|
|
9M29
 
 | |
9M2U
 
 | |
9LZP
 
 | |
9IB0
 
 | |
9IB2
 
 | |
9LVS
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein in complex with S416 | | Descriptor: | 2-[(E)-[[4-(2-chlorophenyl)-1,3-thiazol-2-yl]-methyl-hydrazinylidene]methyl]benzoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fang, Y, Binghao, Z. | | Deposit date: | 2025-02-12 | | Release date: | 2026-03-04 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Cryo-EM structure of the SARS-CoV-2 spike protein(one RBD up state)
To Be Published
|
|
9IB1
 
 | | Cryo-EM focus map of prefusion SARS-CoV-2 spike (RBDs: 1 up & 2 down) bound to RBD-targeting MO176-117 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of antibody MO176-117, Light chain of antibody MO176-117, ... | | Authors: | Schulte, T, Wallden, W, Andrell, J, Ohlin, M. | | Deposit date: | 2025-02-11 | | Release date: | 2026-03-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A stochastic codon deletion generates a SARS-CoV-2-neutralizing antibody
To Be Published
|
|
9IB3
 
 | | Cryo-EM focus map of prefusion SARS-CoV-2 spike (RBDs: 2 up & 1 down) bound to RBD-targeting MO176-117 antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of antibody MO176-117, Light chain of antibody MO176-117, ... | | Authors: | Schulte, T, Wallden, W, Andrell, J, Ohlin, M. | | Deposit date: | 2025-02-11 | | Release date: | 2026-03-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A stochastic codon deletion generates a SARS-CoV-2-neutralizing antibody
To Be Published
|
|
9T74
 
 | |








