6ZH9
 
 | | Ternary complex CR3022 H11-H4 and RBD (SARS-CoV-2) | | Descriptor: | CR3022 Light chain, CR3022 heavy, Nanobody H11-H4, ... | | Authors: | Naismith, J.H, Mikolajek, H, Le Bas, A. | | Deposit date: | 2020-06-21 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7C2Q
 
 | | The crystal structure of COVID-19 main protease in the apo state | | Descriptor: | 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Hu, X.H, Zhou, H, Wang, Q.S, Li, j, Zhang, J. | | Deposit date: | 2020-05-08 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of SARS-CoV-2 main protease in the apo state.
Sci China Life Sci, 64, 2021
|
|
7C2Y
 
 | | The crystal structure of COVID-2019 main protease in the apo state | | Descriptor: | 3C-like proteinase | | Authors: | Zhou, X.L, Zhong, F.L, Lin, C, Zhou, H, Hu, X.H, Wang, Q.S, Li, J, Zhang, J. | | Deposit date: | 2020-05-10 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | COVID-2019 main protease in the apo state
To Be Published
|
|
7C6S
 
 | |
7C6U
 
 | | Crystal structure of SARS-CoV-2 complexed with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Fu, L, Feng, Y, Qi, J, Gao, F.G. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease.
Nat Commun, 11, 2020
|
|
7C8B
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Z-VAD(OMe)-FMK | | Descriptor: | 3C-like proteinase, CHLORIDE ION, Z-VAD(OMe)-FMK | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Yang, S.Y, Lei, J. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the SARS-CoV-2 main protease in complex with Z-VAD(OMe)-FMK
To Be Published
|
|
7C8D
 
 | | Cryo-EM structure of cat ACE2 and SARS-CoV-2 RBD | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S1, ZINC ION | | Authors: | Gao, G.F, Wang, Q.H, Wu, L.l. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Broad host range of SARS-CoV-2 and the molecular basis for SARS-CoV-2 binding to cat ACE2.
Cell Discov, 6, 2020
|
|
7CBT
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Shi, Y, Peng, G. | | Deposit date: | 2020-06-13 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | The preclinical inhibitor GS441524 in combination with GC376 efficaciously inhibited the proliferation of SARS-CoV-2 in the mouse respiratory tract.
Emerg Microbes Infect, 10, 2021
|
|
7CDZ
 
 | |
7CE0
 
 | |
7CJD
 
 | |
7CJM
 
 | | SARS CoV-2 PLpro in complex with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Fu, Z, Huang, H. | | Deposit date: | 2020-07-11 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery.
Nat Commun, 12, 2021
|
|
7CMD
 
 | | Crystal structure of the SARS-CoV-2 PLpro with GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, ZINC ION | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2020-07-27 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease.
Acta Pharm Sin B, 11, 2021
|
|
7CTT
 
 | | Cryo-EM structure of Favipiravir bound to replicating polymerase complex of SARS-CoV-2 in the pre-catalytic state. | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Peng, Q, Peng, R, Shi, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2020-09-02 | | Last modified: | 2021-09-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis of SARS-CoV-2 Polymerase Inhibition by Favipiravir.
Innovation (N Y), 2, 2021
|
|
7JME
 
 | |
7JUN
 
 | |
7JU7
 
 | | The crystal structure of SARS-CoV-2 Main Protease in complex with masitinib | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tan, K, Maltseva, N.I, Welk, L.F, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-08-19 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2.
Science, 373, 2021
|
|
7JYC
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) in Complex with Covalent Inhibitor Narlaprevir | | Descriptor: | (1R,2S,5S)-3-[N-({1-[(tert-butylsulfonyl)methyl]cyclohexyl}carbamoyl)-3-methyl-L-valyl]-N-{(1S)-1-[(1R)-2-(cyclopropylamino)-1-hydroxy-2-oxoethyl]pentyl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Andi, B, Kumaran, D, Kreitler, D.F, Soares, A.S, Shi, W, Jakoncic, J, Fuchs, M.R, Keereetaweep, J, Shanklin, J, McSweeney, S. | | Deposit date: | 2020-08-30 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Hepatitis C virus NS3/4A inhibitors and other drug-like compounds as covalent binders of SARS-CoV-2 main protease.
Sci Rep, 12, 2022
|
|
7A5R
 
 | | Complex of SARS-CoV-2 spike and CR3022 Fab (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab Heavy Chain, CR3022 Fab Light Chain, ... | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-08-21 | | Release date: | 2020-09-16 | | Last modified: | 2020-11-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Antibody-mediated disruption of the SARS-CoV-2 spike glycoprotein.
Nat Commun, 11, 2020
|
|
7A5S
 
 | | Complex of SARS-CoV-2 spike and CR3022 Fab (Homogeneous Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab Heavy Chain, CR3022 Fab Light Chain, ... | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-08-21 | | Release date: | 2020-09-16 | | Last modified: | 2020-11-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Antibody-mediated disruption of the SARS-CoV-2 spike glycoprotein.
Nat Commun, 11, 2020
|
|
7A91
 
 | | Dissociated S1 domain of SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A93
 
 | |
7A94
 
 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A95
 
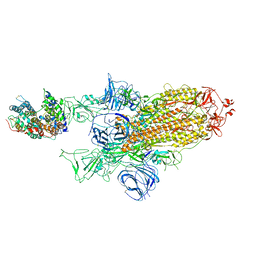 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound and 1 RBD Erect in Clockwise Direction | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A96
 
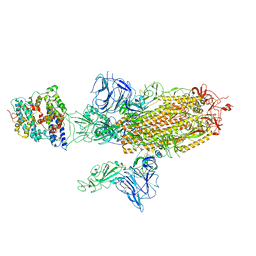 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound and 1 RBD Erect in Anticlockwise Direction | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|







