3L2Y
 
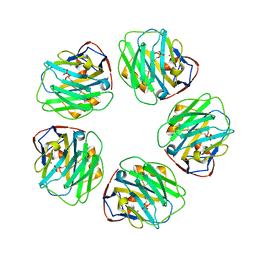 | | The structure of C-reactive protein bound to phosphoethanolamine | | Descriptor: | C-reactive protein, CALCIUM ION, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER | | Authors: | Mikolajek, H, Kolstoe, S.E, Wood, S.P, Pepys, M.B. | | Deposit date: | 2009-12-15 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ligand specificity in the human pentraxins, C-reactive protein and serum amyloid P component.
J.Mol.Recognit., 24, 2011
|
|
5OQG
 
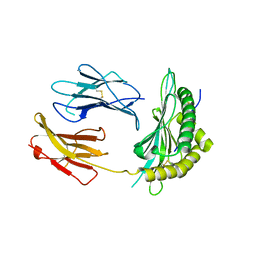 | |
5OQH
 
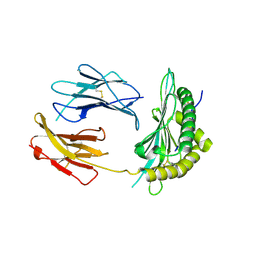 | | Crystal Structure of a disulfide trapped single chain trimer composed of the MHC I heavy chain H-2Kb Y84C K66A mutant, beta-2microglobulin, and ovalbumin-derived peptide | | Descriptor: | Beta-2-microglobulin,H-2 class I histocompatibility antigen, K-B alpha chain | | Authors: | Mikolajek, H, Werner, J.M, Beton, M.E. | | Deposit date: | 2017-08-11 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The partial dissociation of MHC class I-bound peptides exposes their N terminus to trimming by endoplasmic reticulum aminopeptidase 1.
J. Biol. Chem., 293, 2018
|
|
5OQI
 
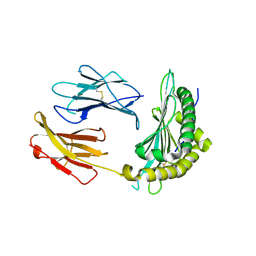 | | Crystal Structure of a disulfide trapped single chain trimer composed of the MHC I heavy chain H-2Kb Y84C E63A mutant, beta-2microglobulin, and ovalbumin-derived peptide | | Descriptor: | Beta-2-microglobulin,H-2 class I histocompatibility antigen, K-B alpha chain | | Authors: | Mikolajek, H, Werner, J.M, Beton, M.E. | | Deposit date: | 2017-08-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The partial dissociation of MHC class I-bound peptides exposes their N terminus to trimming by endoplasmic reticulum aminopeptidase 1.
J. Biol. Chem., 293, 2018
|
|
5OQF
 
 | |
3KQR
 
 | | The structure of serum amyloid p component bound to phosphoethanolamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, ... | | Authors: | Mikolajek, H, Kolstoe, S.E, Wood, S.P, Pepys, M.B. | | Deposit date: | 2009-11-17 | | Release date: | 2010-12-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of ligand specificity in the human pentraxins, C-reactive protein and serum amyloid P component.
J.Mol.Recognit., 24, 2011
|
|
4IHK
 
 | | Crystal structure of the Collagen VI alpha3 N5 domain R1061Q | | Descriptor: | Collagen alpha3(VI) | | Authors: | Mikolajek, H, Becker, A.K.A, Paulsson, M, Wagener, R, Werner, J.M. | | Deposit date: | 2012-12-19 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A structure of a collagen VI VWA domain displays N and C termini at opposite sides of the protein
Structure, 22, 2014
|
|
4IGI
 
 | | Crystal structure of the Collagen VI alpha3 N5 domain | | Descriptor: | Collagen alpha3(VI) | | Authors: | Mikolajek, H, Becker, A.K.A, Paulsson, M, Wagener, R, Werner, J.M. | | Deposit date: | 2012-12-17 | | Release date: | 2013-12-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A structure of a collagen VI VWA domain displays N and C termini at opposite sides of the protein
Structure, 22, 2014
|
|
7Z1A
 
 | | Nanobody H11 and F2 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2 Nanobody, H11 Nanobody, ... | | Authors: | Mikolajek, H, Naismith, J.H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1B
 
 | | Nanobody H11-A10 and F2 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody A10, Nanobody F2, ... | | Authors: | Mikolajek, H, Naismith, J.H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1E
 
 | | Nanobody H11-H4 Q98R H100E bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, H11-H4 Q98R H100E, ... | | Authors: | Mikolajek, H, Naismith, J.H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1C
 
 | | Nanobody H11-B5 and H11-F2 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, Nanobody B5, ... | | Authors: | Mikolajek, H, Naismith, J.H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Z1D
 
 | | Nanobody H11-H6 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, H11-H6 nanobody, ... | | Authors: | Mikolajek, H, Naismith, J.H. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Correlation between the binding affinity and the conformational entropy of nanobody SARS-CoV-2 spike protein complexes.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7ZCK
 
 | | Room temperature crystal structure of PhnD from Synechococcus MITS9220 in complex with phosphate | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Phosphonate ABC type transporter/ substrate binding component | | Authors: | Mikolajek, H, Shah, B.S, Paulsen, I.T, Sandy, J, Sanchez-Weatherby, J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-05-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein-to-structure pipeline for ambient-temperature in situ crystallography at VMXi.
Iucrj, 10, 2023
|
|
7UG8
 
 | | Crystal structure of a solute receptor from Synechococcus CC9311 in complex with alpha-ketovaleric and calcium | | Descriptor: | 1,2-ETHANEDIOL, 2-oxopentanoic acid, CALCIUM ION, ... | | Authors: | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2022-03-24 | | Release date: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Crystal structure of a solute receptor from Synechococcus CC9311 in complex with alpha-ketovaleric and calcium
To Be Published
|
|
7OAY
 
 | | Nanobody F2 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F2 nanobody, Spike protein S1 | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAU
 
 | | Nanobody C5 bound to Kent variant RBD (N501Y) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C5, GLYCEROL, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAQ
 
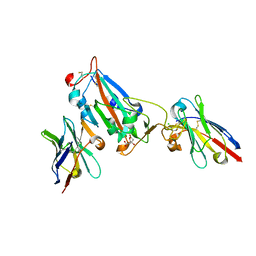 | | Nanobody H3 AND C1 bound to RBD with Kent mutation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-20 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7OAP
 
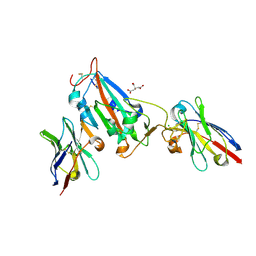 | | Nanobody H3 AND C1 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C1 nanobody, CHLORIDE ION, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
2IZP
 
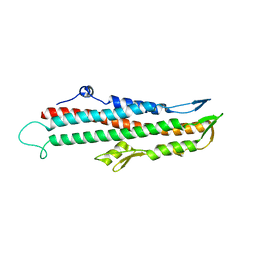 | | BipD - an invasion protein associated with the type-III secretion system of Burkholderia pseudomallei. | | Descriptor: | PUTATIVE MEMBRANE ANTIGEN | | Authors: | Erskine, P.T, Knight, M.J, Ruaux, A, Mikolajek, H, Wong-Fat-Sang, N, Withers, J, Gill, R, Wood, S.P, Wood, M, Fox, G.C, Cooper, J.B. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-06 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High Resolution Structure of Bipd: An Invasion Protein Associated with the Type III Secretion System of Burkholderia Pseudomallei.
J.Mol.Biol., 363, 2006
|
|
6ZH9
 
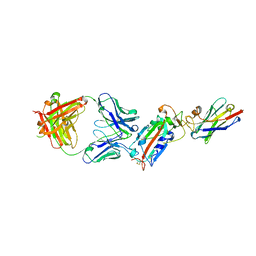 | | Ternary complex CR3022 H11-H4 and RBD (SARS-CoV-2) | | Descriptor: | CR3022 Light chain, CR3022 heavy, Nanobody H11-H4, ... | | Authors: | Naismith, J.H, Mikolajek, H, Le Bas, A. | | Deposit date: | 2020-06-21 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7OAO
 
 | | Nanobody C5 bound to RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, C5 nanobody, ... | | Authors: | Naismith, J.H, Mikolajek, H. | | Deposit date: | 2021-04-19 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19.
Nat Commun, 12, 2021
|
|
7S6G
 
 | | Crystal structure of PhnD from Synechococcus MITS9220 in complex with phosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | Deposit date: | 2021-09-14 | | Release date: | 2021-10-27 | | Last modified: | 2023-07-05 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Marine picocyanobacterial PhnD1 shows specificity for various phosphorus sources but likely represents a constitutive inorganic phosphate transporter.
Isme J, 17, 2023
|
|
8BS8
 
 | | Bovine naive ultralong antibody AbD08 collected at 100K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Heavy chain, Light chain | | Authors: | Clarke, J.D, Douangamath, A, Mikolajek, H, Stuart, D.I, Owens, R.J. | | Deposit date: | 2022-11-24 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Protein-to-structure pipeline for ambient-temperature in situ crystallography at VMXi.
Iucrj, 10, 2023
|
|
8CIF
 
 | | Bovine naive ultralong antibody AbD08 collected at 293K | | Descriptor: | Heavy chain, Light chain | | Authors: | Clarke, J.D, Mikolajek, H, Stuart, D.I, Owens, R.J. | | Deposit date: | 2023-02-09 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Protein-to-structure pipeline for ambient-temperature in situ crystallography at VMXi.
Iucrj, 10, 2023
|
|
