-Search query
-Search result
Showing all 50 items for (author: narita & a)

EMDB-28957: 
Cryo-EM structure of the Agrobacterium T-pilus

PDB-8fai: 
Cryo-EM structure of the Agrobacterium T-pilus
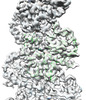
EMDB-33007: 
A CBg-ParM filament with ADP
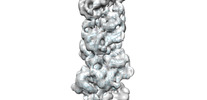
EMDB-33008: 
A CBg-ParM filament with GTP and a short incubation time
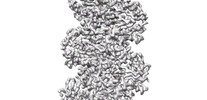
EMDB-33009: 
A CBg-ParM filament with ADP
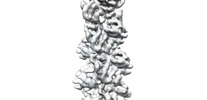
EMDB-33012: 
A CBg-ParM filament with GTP or GDPPi

PDB-7x54: 
A CBg-ParM filament with ADP

PDB-7x55: 
A CBg-ParM filament with GTP and a short incubation time

PDB-7x56: 
A CBg-ParM filament with ADP

PDB-7x59: 
A CBg-ParM filament with GTP or GDPPi

EMDB-14181: 
Human collided disome (di-ribosome) stalled on XBP1 mRNA

PDB-7qvp: 
Human collided disome (di-ribosome) stalled on XBP1 mRNA

EMDB-14153: 
SARS-CoV-2 Spike, C3 symmetry

PDB-7qus: 
SARS-CoV-2 Spike, C3 symmetry

EMDB-14152: 
SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry

EMDB-14154: 
SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C1 symmetry

EMDB-14155: 
SARS-CoV-2 Spike, C1 symmetry

PDB-7qur: 
SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry

EMDB-32629: 
Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution

EMDB-32630: 
Structure and dynamics of Odinarchaeota tubulin and the implications for eukaryotic microtubule evolution

EMDB-30636: 
Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment

PDB-7dce: 
Cryo-EM structure of human XKR8-basigin complex bound to Fab fragment

EMDB-30384: 
Simplified Alpha-Carboxysome, T=3

EMDB-30385: 
Simplified Alpha-Carboxysome, T=4

PDB-7ckb: 
Simplified Alpha-Carboxysome, T=3

PDB-7ckc: 
Simplified Alpha-Carboxysome, T=4

EMDB-9757: 
pCBH ParM filament

EMDB-9758: 
pCBH ParM filament

PDB-6izr: 
Whole structure of a 15-stranded ParM filament from Clostridium botulinum

PDB-6izv: 
Averaged strand structure of a 15-stranded ParM filament from Clostridium botulinum

EMDB-8931: 
afTMEM16 reconstituted in nanodiscs in the absence of Ca2+

EMDB-8948: 
PDB: afTMEM16 reconstituted in nanodiscs in the presence of Ca2+

EMDB-8959: 
afTMEM16 reconstituted in nanodiscs in the presence of Ca2+ and ceramide 24:0

PDB-6dz7: 
afTMEM16 reconstituted in nanodiscs in the absence of Ca2+

PDB-6e0h: 
PDB: afTMEM16 reconstituted in nanodiscs in the presence of Ca2+

PDB-6e1o: 
afTMEM16 reconstituted in nanodiscs in the presence of Ca2+ and ceramide 24:0

EMDB-6989: 
Inner capsid of Rice Dwarf Virus with T-trimers (Full particle)

EMDB-6990: 
Inner capsid of Rice Dwarf Virus with T-trimers (empty particle)

EMDB-9618: 
Localization of FAP70 N-terminus on Chlamydomonas axoneme

EMDB-6712: 
Inner capsid of Rice Dwarf Virus

EMDB-6713: 
Inner capsid of Rice Dwarf Virus with T-trimers

EMDB-6844: 
Cofilin decorated actin filament

PDB-5yu8: 
Cofilin decorated actin filament

EMDB-2716: 
Dynactin 3D structure: Implications for assembly and dynein binding

EMDB-2068: 
Alp12 filament structure

PDB-4apw: 
Alp12 filament structure

PDB-2y83: 
Actin filament pointed end
EMDB-1872: 
Actin filament pointed end

EMDB-1470: 
Molecular structure of the ParM polymer and the mechanism leading to its nucleotide-driven dynamic instability

PDB-2zhc: 
ParM filament
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
