+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2y83 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Actin filament pointed end | ||||||
 Components Components | ACTIN, ALPHA SKELETAL MUSCLE | ||||||
 Keywords Keywords | CONTRACTILE PROTEIN / CYTOSKELETON / CELL MOTILITY | ||||||
| Function / homology |  Function and homology information Function and homology informationcytoskeletal motor activator activity / myosin heavy chain binding / tropomyosin binding / actin filament bundle / troponin I binding / filamentous actin / mesenchyme migration / skeletal muscle myofibril / actin filament bundle assembly / striated muscle thin filament ...cytoskeletal motor activator activity / myosin heavy chain binding / tropomyosin binding / actin filament bundle / troponin I binding / filamentous actin / mesenchyme migration / skeletal muscle myofibril / actin filament bundle assembly / striated muscle thin filament / skeletal muscle thin filament assembly / actin monomer binding / skeletal muscle fiber development / stress fiber / titin binding / actin filament polymerization / actin filament / filopodium / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding / lamellipodium / cell body / protein domain specific binding / hydrolase activity / calcium ion binding / positive regulation of gene expression / magnesium ion binding / ATP binding / identical protein binding / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 22.9 Å | ||||||
 Authors Authors | Narita, A. / Oda, T. / Maeda, Y. | ||||||
 Citation Citation |  Journal: EMBO J / Year: 2011 Journal: EMBO J / Year: 2011Title: Structural basis for the slow dynamics of the actin filament pointed end. Authors: Akihiro Narita / Toshiro Oda / Yuichiro Maéda /  Abstract: The actin filament has clear polarity where one end, the pointed end, has a much slower polymerization and depolymerization rate than the other end, the barbed end. This intrinsic difference of the ...The actin filament has clear polarity where one end, the pointed end, has a much slower polymerization and depolymerization rate than the other end, the barbed end. This intrinsic difference of the ends significantly affects all actin dynamics in the cell, which has central roles in a wide spectrum of cellular functions. The detailed mechanism underlying this difference has remained elusive, because high-resolution structures of the filament ends have not been available. Here, we present the structure of the actin filament pointed end obtained using a single particle analysis of cryo-electron micrographs. We determined that the terminal pointed end subunit is tilted towards the penultimate subunit, allowing specific and extra loop-to-loop inter-strand contacts between the two end subunits, which is not possible in other parts of the filament. These specific contacts prevent the end subunit from dissociating. For elongation, the loop-to-loop contacts also inhibit the incorporation of another actin monomer at the pointed end. These observations are likely to account for the less dynamic pointed end. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2y83.cif.gz 2y83.cif.gz | 685.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2y83.ent.gz pdb2y83.ent.gz | 566.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2y83.json.gz 2y83.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/y8/2y83 https://data.pdbj.org/pub/pdb/validation_reports/y8/2y83 ftp://data.pdbj.org/pub/pdb/validation_reports/y8/2y83 ftp://data.pdbj.org/pub/pdb/validation_reports/y8/2y83 | HTTPS FTP |
|---|
-Related structure data
| Related structure data | 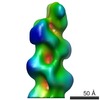 1872MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
| #1: Protein | Mass: 41862.613 Da / Num. of mol.: 6 / Source method: isolated from a natural source / Source: (natural)  #2: Chemical | ChemComp-ADP / #3: Chemical | ChemComp-CA / |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: FILAMENT / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: RABBIT SKELETAL ACTIN / Type: COMPLEX |
|---|---|
| Buffer solution | Name: 50 MM NACL, 10 MM SODIUM PHOSPHATE BUFFER PH 7.4 3 MM MGCL2, 0.005% (W/V) NAN3, 0.7 MM DTT. pH: 7.4 Details: 50 MM NACL, 10 MM SODIUM PHOSPHATE BUFFER PH 7.4 3 MM MGCL2, 0.005% (W/V) NAN3, 0.7 MM DTT. |
| Specimen | Conc.: 0.05 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Details: HOLEY CARBON |
| Vitrification | Cryogen name: ETHANE Details: VITRIFICATION 1 - CRYOGEN- ETHANE, HUMIDITY- 90%, TEMPERATURE- 4 DEGREES CELSIUS. METHOD- BLOT FOR 3 SECONDS BEFORE PLUNGING. |
- Electron microscopy imaging
Electron microscopy imaging
| Microscopy | Model: JEOL 3200FSC |
|---|---|
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 40000 X / Nominal defocus max: 8000 nm / Nominal defocus min: 5000 nm |
| Specimen holder | Temperature: 4 K |
| Image recording | Electron dose: 34 e/Å2 / Film or detector model: KODAK SO-163 FILM |
| Image scans | Num. digital images: 142 |
- Processing
Processing
| EM software | Name: EOS / Category: 3D reconstruction | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Details: PHASE AND AMPLITUDE | ||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||
| 3D reconstruction | Method: SINGLE PARTICLE ANALYSIS / Resolution: 22.9 Å / Num. of particles: 714 / Nominal pixel size: 3.4125 Å / Actual pixel size: 3.4125 Å Details: SUBMISSION BASED ON EXPERIMENTAL DATA FROM EMDB EMD-1872. Symmetry type: POINT | ||||||||||||
| Atomic model building | Protocol: FLEXIBLE FIT / Space: REAL Details: METHOD--RIGID BODY AND MOLECULAR DYNAMICS REFINEMENT PROTOCOL--EM | ||||||||||||
| Atomic model building | PDB-ID: 2ZWH Accession code: 2ZWH / Source name: PDB / Type: experimental model | ||||||||||||
| Refinement | Highest resolution: 22.9 Å | ||||||||||||
| Refinement step | Cycle: LAST / Highest resolution: 22.9 Å
|
 Movie
Movie Controller
Controller




































































 PDBj
PDBj







