+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8931 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | afTMEM16 reconstituted in nanodiscs in the absence of Ca2+ | |||||||||
 Map data Map data | afTMEM16 reconstituted in nanodiscs in the absence of Ca2+ | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | scramblase / Ca2+-activated / membrane-reorganization / LIPID TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationphospholipid scramblase activity / cortical endoplasmic reticulum / phospholipid translocation / chloride channel activity / voltage-gated calcium channel activity / chloride transmembrane transport / monoatomic ion transmembrane transport / membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
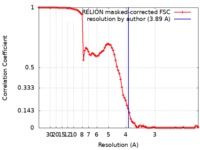
| Method | single particle reconstruction / cryo EM / Resolution: 3.89 Å | |||||||||
 Authors Authors | Falzone ME / Accardi A | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2019 Journal: Elife / Year: 2019Title: Structural basis of Ca-dependent activation and lipid transport by a TMEM16 scramblase. Authors: Maria E Falzone / Jan Rheinberger / Byoung-Cheol Lee / Thasin Peyear / Linda Sasset / Ashleigh M Raczkowski / Edward T Eng / Annarita Di Lorenzo / Olaf S Andersen / Crina M Nimigean / Alessio Accardi /   Abstract: The lipid distribution of plasma membranes of eukaryotic cells is asymmetric and phospholipid scramblases disrupt this asymmetry by mediating the rapid, nonselective transport of lipids down their ...The lipid distribution of plasma membranes of eukaryotic cells is asymmetric and phospholipid scramblases disrupt this asymmetry by mediating the rapid, nonselective transport of lipids down their concentration gradients. As a result, phosphatidylserine is exposed to the outer leaflet of membrane, an important step in extracellular signaling networks controlling processes such as apoptosis, blood coagulation, membrane fusion and repair. Several TMEM16 family members have been identified as Ca-activated scramblases, but the mechanisms underlying their Ca-dependent gating and their effects on the surrounding lipid bilayer remain poorly understood. Here, we describe three high-resolution cryo-electron microscopy structures of a fungal scramblase from , afTMEM16, reconstituted in lipid nanodiscs. These structures reveal that Ca-dependent activation of the scramblase entails global rearrangement of the transmembrane and cytosolic domains. These structures, together with functional experiments, suggest that activation of the protein thins the membrane near the transport pathway to facilitate rapid transbilayer lipid movement. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8931.map.gz emd_8931.map.gz | 57.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8931-v30.xml emd-8931-v30.xml emd-8931.xml emd-8931.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8931_fsc.xml emd_8931_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_8931.png emd_8931.png | 51.8 KB | ||
| Filedesc metadata |  emd-8931.cif.gz emd-8931.cif.gz | 6 KB | ||
| Others |  emd_8931_additional.map.gz emd_8931_additional.map.gz | 50.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8931 http://ftp.pdbj.org/pub/emdb/structures/EMD-8931 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8931 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8931 | HTTPS FTP |
-Related structure data
| Related structure data |  6dz7MC  8948C  8959C  6e0hC  6e1oC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8931.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8931.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | afTMEM16 reconstituted in nanodiscs in the absence of Ca2+ | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0961 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
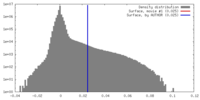
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: unmasked C1 map containing the nanodisc density used...
| File | emd_8931_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unmasked C1 map containing the nanodisc density used for membrane analysis | ||||||||||||
| Projections & Slices |
| ||||||||||||
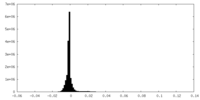
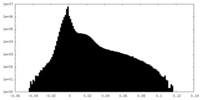
| Density Histograms |
- Sample components
Sample components
-Entire : afTMEM16 reconstituted in nanodiscs in the absence of Ca2+
| Entire | Name: afTMEM16 reconstituted in nanodiscs in the absence of Ca2+ |
|---|---|
| Components |
|
-Supramolecule #1: afTMEM16 reconstituted in nanodiscs in the absence of Ca2+
| Supramolecule | Name: afTMEM16 reconstituted in nanodiscs in the absence of Ca2+ type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 168 kDa/nm |
-Macromolecule #1: Plasma membrane channel protein (Aqy1), putative
| Macromolecule | Name: Plasma membrane channel protein (Aqy1), putative / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Strain: ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100 |
| Molecular weight | Theoretical: 84.616859 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAFNPAPKAV QENHHVDYVI RFNYGDIDTP EAIKKFEVLL LELSEVGLQT EVRQGDENSL FVFVRAASKK KLKRAVYQSR VRDWLYGVR NTEPEPASSA KPQSEAERLL VIYHLITVPK AEGGAGITPR HGEWKNVDAI FPLHDEETNR QCMREWSKKT F LSTEDLDR ...String: MAFNPAPKAV QENHHVDYVI RFNYGDIDTP EAIKKFEVLL LELSEVGLQT EVRQGDENSL FVFVRAASKK KLKRAVYQSR VRDWLYGVR NTEPEPASSA KPQSEAERLL VIYHLITVPK AEGGAGITPR HGEWKNVDAI FPLHDEETNR QCMREWSKKT F LSTEDLDR IRNTFGEHVG FYFAFLQSYF RFLMFPAAFG FSCWLLLGSF SIIYTVVNCL WCIVFIEYWK RQEEDLSCRW QT KGVSAVH EKRAEFKPEK EIRDESTGEV RGVFPATKRM YRQLLQVPFA LLAAVALGAI IATCFAIEIF ISEVYNGPLK GYL VFIPTI LVSALIPTMS AVLLTVATKL NDYENYETQD AYKVALTQKI FVVNFITSYL PIILTAFVYV PFASRIVPYL DVFH LTVRP FVSKEHAIKA RTEFSINPDR LRKQVIYFTV TAQIVGFALE TIVPFVKQRV FREYKEYTKK QHAKAEPGNG AGEKK TVSL GDDEDEARFL TRVRNEAELE DYDVTDDLRE MCIQFGYLAL FSPVWPLVPV SFLINNWVEL RSDFFKICVE CKRPWP QRA DTIGPWLDSL GFLSWVGSIT SSALVYMFSN GHEGPNGEPT TIRCWALLLT IFFSEHLYLI VRYAVRSALA KLEPPNT RR ERIERFMMRK RYLDTVLSAE SDDDADEVKG VVSSIPPSEI TRESLEQDAR DWSKQGTDPT ERFWMRQRGW KESAEVGL S LITKAKGDET KKQQ UniProtKB: Plasma membrane channel protein (Aqy1), putative |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 7 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 288 K / Instrument: FEI VITROBOT MARK IV |
| Details | afTMEM16 reconstituted in nanodiscs in the absence of Ca2+ |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Average electron dose: 62.61 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)