+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6989 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Inner capsid of Rice Dwarf Virus with T-trimers (Full particle) | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |   Rice dwarf virus Rice dwarf virus | ||||||||||||
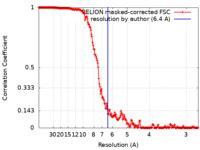
| Method | single particle reconstruction / cryo EM / Resolution: 6.4 Å | ||||||||||||
 Authors Authors | Nakamichi Y / Miyazaki N / Tsutsumi K / Higashiura A / Narita H / Murata K / Nakagawa A | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Structure / Year: 2019 Journal: Structure / Year: 2019Title: An Assembly Intermediate Structure of Rice Dwarf Virus Reveals a Hierarchical Outer Capsid Shell Assembly Mechanism. Authors: Yusuke Nakamichi / Naoyuki Miyazaki / Kenta Tsutsumi / Akifumi Higashiura / Hirotaka Narita / Kazuyoshi Murata / Atsushi Nakagawa /  Abstract: Nearly all viruses of the Reoviridae family possess a multi-layered capsid consisting of an inner layer with icosahedral T = 1 symmetry and a second-outer layer (composed of 260 copies of a trimeric ...Nearly all viruses of the Reoviridae family possess a multi-layered capsid consisting of an inner layer with icosahedral T = 1 symmetry and a second-outer layer (composed of 260 copies of a trimeric protein) exhibiting icosahedral T = 13 symmetry. Here we describe the construction and structural evaluation of an assembly intermediate of the Rice dwarf virus of the family Reoviridae stalled at the second capsid layer via targeted disruption of the trimer-trimer interaction interface in the second-layer capsid protein. Structural determination was performed by conventional and Zernike/Volta phase-contrast cryoelectron microscopy. The assembly defect second-layer capsid trimers bound exclusively to the outer surface of the innermost capsid layer at the icosahedral 3-fold axis. Furthermore, the second-layer assembly could not proceed without specific inter-trimer interactions. Our results suggest that the correct assembly pathway for second-layer capsid formation is highly controlled at the inter-layer and inter-trimer interactions. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6989.map.gz emd_6989.map.gz | 112.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6989-v30.xml emd-6989-v30.xml emd-6989.xml emd-6989.xml | 11.8 KB 11.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6989_fsc.xml emd_6989_fsc.xml | 19.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_6989.png emd_6989.png | 263.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6989 http://ftp.pdbj.org/pub/emdb/structures/EMD-6989 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6989 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6989 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6989.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6989.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
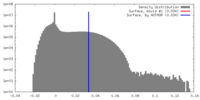
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Rice dwarf virus
| Entire | Name:   Rice dwarf virus Rice dwarf virus |
|---|---|
| Components |
|
-Supramolecule #1: Rice dwarf virus
| Supramolecule | Name: Rice dwarf virus / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 10991 / Sci species name: Rice dwarf virus / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 6 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: MOLYBDENUM / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 90.0 K / Max: 95.0 K |
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Spherical aberration corrector: CEOS Cs-corrector |
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Detector mode: INTEGRATING / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Digitization - Sampling interval: 14.0 µm / Digitization - Frames/image: 2-30 / Number grids imaged: 1 / Number real images: 2804 / Average exposure time: 2.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.1 mm / Nominal defocus max: -1.2 µm / Nominal defocus min: -1.0 µm / Nominal magnification: 47000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)