7NNS
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Momelotinib | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type I, Momelotinib, ... | | Authors: | Williams, E, Chen, Z, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2021-02-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound Momelotinib
To Be Published
|
|
9BCZ
 
 | | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Edwards, M.M, Dong, A, Theo-Emegano, N, Seitova, A, Loppnau, P, Leung, R, Li, H, Ilyassov, O, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-10 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Chicken 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase zeta-1 (PLCZ1) in complex with calcium and phosphorylated threonine
To be published
|
|
7RUO
 
 | | Crystal structure of human UTP15 | | Descriptor: | U3 small nucleolar RNA-associated protein 15 homolog, UNKNOWN ATOM OR ION | | Authors: | Dehghani-Tafti, S, Dong, A, Zeng, H, Hutchinson, A, Seitova, A, Loppnau, P, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-08-17 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of human UTP15
To Be Published
|
|
1L1S
 
 | | Structure of Protein of Unknown Function MTH1491 from Methanobacterium thermoautotrophicum | | Descriptor: | hypothetical protein MTH1491 | | Authors: | Christendat, D, Saridakis, V, Kim, Y, Kumar, P.A, Xu, X, Semesi, A, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-19 | | Release date: | 2002-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of hypothetical protein MTH1491 from Methanobacterium thermoautotrophicum.
Protein Sci., 11, 2002
|
|
9MJG
 
 | | Crystal structure of HAT1 in complex with XS380871 | | Descriptor: | (4S,7R)-4-(3-ethoxy-4-hydroxy-5-nitrophenyl)-7-(4-fluorophenyl)-4,6,7,8-tetrahydroquinoline-2,5(1H,3H)-dione, Histone acetyltransferase type B catalytic subunit | | Authors: | Zeng, H, Li, F, Wang, X, Sun, J, Dong, A, Peng, H, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-12-15 | | Release date: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of HAT1 in complex with XS380871
To be published
|
|
9MK7
 
 | | Crystal structure of the WDR domain of WDR91 in complex with DR3634 | | Descriptor: | (1R)-1-({(6M)-2-(methylamino)-6-[3-(pyrrolidin-1-yl)phenyl]pyrimidin-4-yl}amino)-2,3-dihydro-1H-indene-4-carbonitrile, UNKNOWN ATOM OR ION, WD repeat-containing protein 91 | | Authors: | Zeng, H, Ahmad, H, Dong, A, Seitova, A, Owen, D, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-12-16 | | Release date: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of the WDR domain of WDR91 in complex with DR3634
To be published
|
|
6EDF
 
 | | Fragment of a tyrosine-protein kinase | | Descriptor: | FYN, GLYCEROL, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Huang, H, Sochirca, I, Liu, K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Sidhu, S.S, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-08-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fragment of a tyrosine-protein kinase
To be Published
|
|
6FIC
 
 | | Bivalent Inhibitor UNC4512 Bound to the TAF1 Bromodomain Tandem | | Descriptor: | 3-azanyl-2-[3-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[2-[[3-azanyl-1-[[2-[[3-methyl-6-[4-methyl-3-(methylsulfonyl-$l^{2}-azanyl)cyclohexa-1,3,5-trien-1-yl]-[1,2,4]triazolo[4,3-b]pyridazin-8-yl]-$l^{2}-azanyl]-2-oxidanylidene-ethyl]amino]-1-oxidanylidene-propan-2-yl]amino]-2-oxidanylidene-ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]propanoylamino]-~{N}-[3-[[3-methyl-6-[4-methyl-3-(methylsulfonylamino)phenyl]-[1,2,4]triazolo[4,3-b]pyridazin-8-yl]amino]-3-oxidanylidene-propyl]propanamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Mathea, S, Suh, J.L, Salah, E, Tallant, C, Siejka, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, James, L.I, Frye, S.V, Knapp, S. | | Deposit date: | 2018-01-17 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Bivalent Inhibitor UNC4512 Bound to the TAF1 Bromodomain Tandem
To Be Published
|
|
5JUW
 
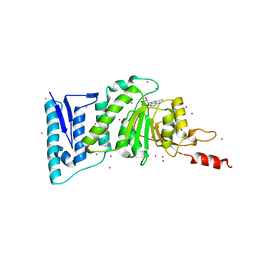 | | complex of Dot1l with SS148 | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Li, Y, Spurr, S.S, Bayle, E.D, Fish, P.V, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-10 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Complex of Dot1l with SS148
To Be Published
|
|
5K29
 
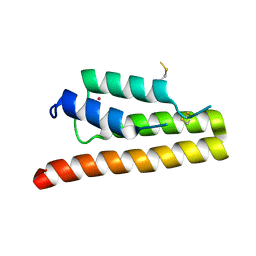 | | Trypanosoma brucei bromodomain BDF5 (Tb427tmp.01.5000) | | Descriptor: | UNKNOWN ATOM OR ION, uncharacterized protein BDF5 | | Authors: | Lin, Y.H, Tempel, W, Walker, J.R, Loppnau, P, Amani, M, Hou, C.F.D, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-05-18 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trypanosoma brucei bromodomain BDF5 (Tb427tmp.01.5000)
To Be Published
|
|
9AY9
 
 | | Co-crystal structure of human PRMT9 in complex with MRK-990 inhibitor | | Descriptor: | 7-[5-S-(4-{[(2-tert-butylpyridin-3-yl)methyl]amino}butyl)-5-thio-beta-D-ribofuranosyl]-5-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Protein arginine N-methyltransferase 9, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Li, Y, Gao, Y.D, Schneider, S, Siliphaivanh, P, Sloman, D, Nicholson, B, Fischer, C, Hicks, J, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-07 | | Release date: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Co-crystal structure of human PRMT9 in complex with MRK-990 inhibitor
To be published
|
|
9AVA
 
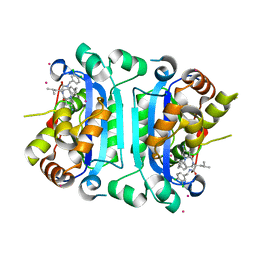 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | (2R)-2-[(5R,6S,8R,9aS)-8-amino-1-oxo-5-(2-phenylethyl)hexahydro-1H-pyrrolo[1,2-a][1,4]diazepin-2(3H)-yl]-N-[(3,4-dichlorophenyl)methyl]-4-methylpentanamide, POTASSIUM ION, Three-prime repair exonuclease 1, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Xu, J, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-03-01 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
4UYI
 
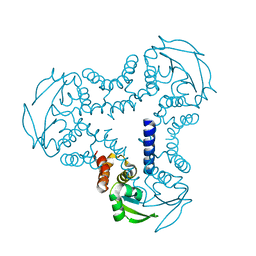 | | Crystal structure of the BTB domain of human SLX4 (BTBD12) | | Descriptor: | STRUCTURE-SPECIFIC ENDONUCLEASE SUBUNIT SLX4 | | Authors: | Pinkas, D.M, Sanvitale, C.E, Strain-Damerell, C, Fairhead, M, Wang, D, Tallant, C, Cooper, C.D.O, Sorrell, F.J, Kopec, J, Chaikuad, A, Fitzpatrick, F, Pike, A.C.W, Hozjan, V, Ying, Z, Roos, A.K, Savitsky, P, Bradley, A, Nowak, R, Filippakopoulos, P, Krojer, T, Burgess-Brown, N.A, Marsden, B.D, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-09-01 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of the Btb Domain of Human Slx4 (Btbd12)
To be Published
|
|
5CZG
 
 | | Crystal Structure Analysis of hypothetical bromodomain Tb427.10.7420 from Trypanosoma brucei in complex with bromosporine | | Descriptor: | Bromosporine, Hypothetical Bromodomain, SODIUM ION, ... | | Authors: | Jiang, D.Q, Tempel, W, Loppnau, P, Graslund, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Amani, M, Hou, C.F.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-31 | | Release date: | 2015-08-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Crystal Structure Analysis of hypothetical bromodomain from Trypanosoma brucei
to be published
|
|
9O0X
 
 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-({N-[(2,3-dichlorophenyl)methyl]-3,5-difluorobenzamido}methyl)-3-(methylamino)pyridine-2-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-04-03 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
9QOB
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound 4 | | Descriptor: | (5-phenyl-[1,3]thiazolo[2,3-c][1,2,4]triazol-3-yl)methanol, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F. | | Deposit date: | 2025-03-25 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound 2
To Be Published
|
|
9O0W
 
 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-({N-[(2,3-dichlorophenyl)methyl]-3,5-difluorobenzamido}methyl)pyridine-2-carboxylic acid, MAGNESIUM ION, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-04-03 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
9O0Y
 
 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-[(2,3-dichlorophenyl)methyl]-3,5-difluoro-N-[(pyridin-3-yl)methyl]benzamide, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-04-03 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
9NIU
 
 | | Co-crystal structure of FABP7 in complex with PFO3TDA | | Descriptor: | 1,2-ETHANEDIOL, Fatty acid-binding protein, brain, ... | | Authors: | Yang, D, Liu, J, Zeng, H, Dong, A, Arrowsmith, C.H, Edwards, A.M, Peng, H, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-02-26 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Co-crystal structure of FABP7 in complex with PFO3TDA
To be published
|
|
9QNV
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-5-phenyl-[1,3]thiazolo[2,3-c][1,2,4]triazole, Bromodomain-containing protein 4, ... | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-03-25 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.226 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound 2
To Be Published
|
|
7NSG
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B | | Descriptor: | (+)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, (-)-3-Hydroxy-4,5,6,6a-tetrahydro-3aH-pyrrolo[3,4-d]isoxazole-6-carboxylic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S.M.M, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-05 | | Release date: | 2022-03-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with HIP-B
TO BE PUBLISHED
|
|
7B9W
 
 | | Crystal structure of MurE from E.coli in complex with Z757284380 | | Descriptor: | 1,2-ETHANEDIOL, 3-Chloro-N-[2-(2,4-dioxo-1,3-thiazolidin-3-yl)ethyl]benzamide, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate-2,6-diaminopimelate ligase | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-14 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
7BBP
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with H4K5acK8ac | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Histone H4, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with H4K5acK8ac
To Be Published
|
|
7BBO
 
 | | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystal Structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in space group P21212
To Be Published
|
|
4LDG
 
 | | Crystal Structure of CpSET8 from Cryptosporidium, cgd4_370 | | Descriptor: | MALONATE ION, Protein with a SET domain within carboxy region | | Authors: | Wernimont, A.K, Tempel, W, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-24 | | Release date: | 2013-07-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of CpSET8 from Cryptosporidium, cgd4_370
To be Published
|
|
