1QJT
 
 | | SOLUTION STRUCTURE OF THE APO EH1 DOMAIN OF MOUSE EPIDERMAL GROWTH FACTOR RECEPTOR SUBSTRATE 15, EPS15 | | Descriptor: | EPIDERMAL GROWTH FACTOR RECEPTOR SUBSTRATE SUBSTRATE 15, EPS15 | | Authors: | Whitehead, B, Tessari, M, Carotenuto, A, van Bergen en Henegouwen, P.M, Vuister, G.W. | | Deposit date: | 1999-07-02 | | Release date: | 2000-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Eh1 Domain of Eps15 is Structurally Classified as a Member of the S100 Subclass of EF-Hand Containing Proteins
Biochemistry, 38, 1999
|
|
1GWZ
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF THE PROTEIN TYROSINE PHOSPHATASE SHP-1 | | Descriptor: | SHP-1 | | Authors: | Yang, J, Liang, X, Niu, T, Meng, W, Zhao, Z, Zhou, G.W. | | Deposit date: | 1998-08-22 | | Release date: | 1999-08-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the catalytic domain of protein-tyrosine phosphatase SHP-1.
J.Biol.Chem., 273, 1998
|
|
5OEO
 
 | | Solution structure of the complex of TRPV5(655-725) with a Calmodulin E32Q/E68Q double mutant | | Descriptor: | CALCIUM ION, Calmodulin-1, Transient receptor potential cation channel subfamily V member 5 | | Authors: | Vuister, G.W, Bokhovchuk, F.M, Bate, N, Kovalevskaya, N, Goult, B.T, Spronk, C.A.E.M. | | Deposit date: | 2017-07-09 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structural Basis of Calcium-Dependent Inactivation of the Transient Receptor Potential Vanilloid 5 Channel.
Biochemistry, 57, 2018
|
|
5T1A
 
 | | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{R})-1-(4-chloranyl-2-fluoranyl-phenyl)-2-cyclohexyl-3-ethanoyl-4-oxidanyl-2~{H}-pyrrol-5-one, (3S)-1-{(1S,2R,4R)-4-[methyl(propan-2-yl)amino]-2-propylcyclohexyl}-3-{[6-(trifluoromethyl)quinazolin-4-yl]amino}pyrrolidin-2-one, ... | | Authors: | Zheng, Y, Qin, L, Ortiz Zacarias, N.V, de Vries, H, Han, G.W, Gustavsson, M, Dabros, M, Zhao, C, Cherney, R.J, Carter, P, Stamos, D, Abagyan, R, Cherezov, V, Stevens, R.C, IJzerman, A.P, Heitman, L.H, Tebben, A, Kufareva, I, Handel, T.M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists.
Nature, 540, 2016
|
|
5TGZ
 
 | | Crystal Structure of the Human Cannabinoid Receptor CB1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-[4-[2-(2,4-dichlorophenyl)-4-methyl-5-(piperidin-1-ylcarbamoyl)pyrazol-3-yl]phenyl]but-3-ynyl nitrate, Cannabinoid receptor 1,Flavodoxin,Cannabinoid receptor 1, ... | | Authors: | Hua, T, Vemuri, K, Pu, M, Qu, L, Han, G.W, Wu, Y, Zhao, S, Shui, W, Li, S, Korde, A, Laprairie, R.B, Stahl, E.L, Ho, J.H, Zvonok, N, Zhou, H, Kufareva, I, Wu, B, Zhao, Q, Hanson, M.A, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2016-09-28 | | Release date: | 2016-11-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Human Cannabinoid Receptor CB1.
Cell, 167, 2016
|
|
2XEZ
 
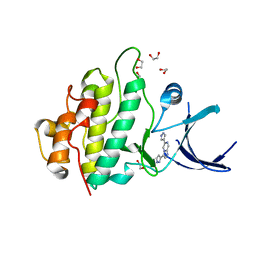 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 6-(1H-PYRAZOL-3-YL)-3-(1H-PYRAZOL-4-YL)IMIDAZO[1,2-A]PYRAZINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, McHardy, T, Klair, S, Boxall, K, Fisher, M, Cherry, M, Allen, C.E, Addison, G.J, Ellard, J, Aherne, G.W, Westwood, I.M, vanMontfort, R, Garrett, M.D, Reader, J.C, Collins, I. | | Deposit date: | 2010-05-19 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design and Evaluation of 3,6-Di(Hetero)Aryl Imidazo[1,2-A]Pyrazines as Inhibitors of Checkpoint and Other Kinases.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1GXM
 
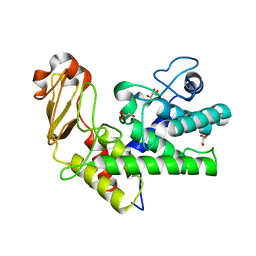 | | Family 10 polysaccharide lyase from Cellvibrio cellulosa | | Descriptor: | GLYCEROL, PECTATE LYASE | | Authors: | Charnock, S.J, Brown, I.E, Turkenburg, J.P, Black, G.W, Davies, G.J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Convergent Evolution Sheds Light on the Anti-Beta-Elimination Mechanism Common to Family 1 and 10 Polysaccharide Lyases
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GXO
 
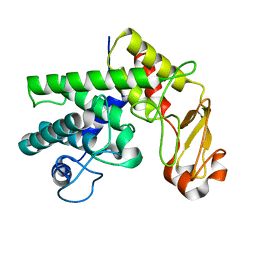 | | Mutant D189A of Family 10 polysaccharide lyase from Cellvibrio cellulosa in complex with trigalaturonic acid | | Descriptor: | CALCIUM ION, PECTATE LYASE, alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid-(1-4)-alpha-D-galactopyranuronic acid | | Authors: | Charnock, S.J, Brown, I.E, Turkenburg, J.P, Black, G.W, Davies, G.J. | | Deposit date: | 2002-04-08 | | Release date: | 2002-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Convergent Evolution Sheds Light on the Anti-Beta-Elimination Mechanism Common to Family 1 and 10 Polysaccharide Lyases
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6W25
 
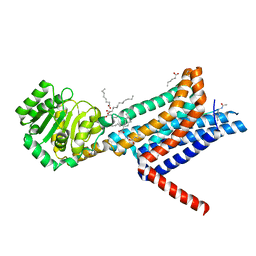 | | Crystal structure of the Melanocortin-4 Receptor (MC4R) in complex with SHU9119 | | Descriptor: | CALCIUM ION, Melanocortin receptor 4,GlgA glycogen synthase,Melanocortin receptor 4, OLEIC ACID, ... | | Authors: | Yu, J, Gimenez, L.E, Hernandez, C.C, Wu, Y, Wein, A.H, Han, G.W, McClary, K, Mittal, S.R, Burdsall, K, Stauch, B, Wu, L, Stevens, S.N, Peisley, A, Williams, S.Y, Chen, V, Millhauser, G.L, Zhao, S, Cone, R.D, Stevens, R.C. | | Deposit date: | 2020-03-04 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Determination of the melanocortin-4 receptor structure identifies Ca2+as a cofactor for ligand binding.
Science, 368, 2020
|
|
6VJM
 
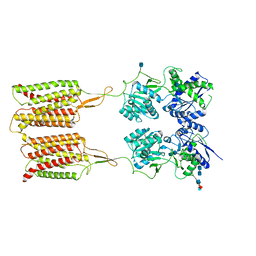 | | Human metabotropic GABA(B) receptor in its apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shaye, H, Han, G.W, Gati, C, Cherezov, V. | | Deposit date: | 2020-01-16 | | Release date: | 2020-06-10 | | Last modified: | 2020-08-26 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural basis of the activation of a metabotropic GABA receptor.
Nature, 584, 2020
|
|
5XF1
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
2A7Y
 
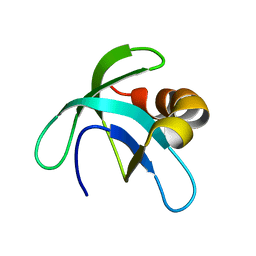 | | Solution Structure of the Conserved Hypothetical Protein Rv2302 from the Bacterium Mycobacterium tuberculosis | | Descriptor: | Hypothetical protein Rv2302/MT2359 | | Authors: | Buchko, G.W, Kim, C.-Y, Terwilliger, T.C, Kennedy, M.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-06 | | Release date: | 2005-08-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the conserved hypothetical protein Rv2302 from Mycobacterium tuberculosis.
J.Bacteriol., 188, 2006
|
|
5XRA
 
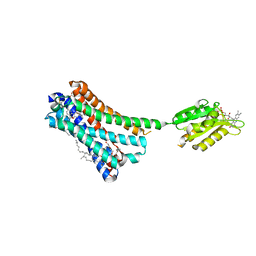 | | Crystal structure of the human CB1 in complex with agonist AM11542 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6aR,10aR)-3-(8-bromanyl-2-methyl-octan-2-yl)-6,6,9-trimethyl-6a,7,10,10a-tetrahydrobenzo[c]chromen-1-ol, CHOLESTEROL, ... | | Authors: | Hua, T, Vemuri, K, Nikas, P.S, Laprairie, R.B, Wu, Y, Qu, L, Pu, M, Korde, A, Shan, J, Ho, J.H, Han, G.W, Ding, K, Li, X, Liu, H, Hanson, M.A, Zhao, S, Bohn, L.M, Makriyannis, A, Stevens, R.C, Liu, Z.J. | | Deposit date: | 2017-06-08 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of agonist-bound human cannabinoid receptor CB1
Nature, 547, 2017
|
|
5X8I
 
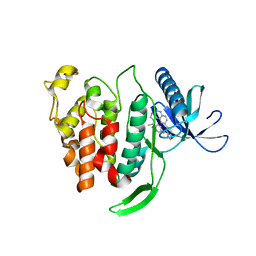 | | Crystal structure of human CLK1 in complex with compound 25 | | Descriptor: | 5-[1-[(1S)-1-(4-fluorophenyl)ethyl]-[1,2,3]triazolo[4,5-c]quinolin-8-yl]-1,3-benzoxazole, Dual specificity protein kinase CLK1 | | Authors: | Sun, Q.Z, Lin, G.F, Li, L.L, Jin, X.T, Huang, L.Y, Zhang, G, Wei, Y.Q, Lu, G.W, Yang, S.Y. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Cdc2-Like Kinase 1 (CLK1) as a New Class of Autophagy Inducers
J. Med. Chem., 60, 2017
|
|
2B3R
 
 | | Crystal structure of the C2 domain of class II phosphatidylinositide 3-kinase C2 | | Descriptor: | Phosphatidylinositol-4-phosphate 3-kinase C2 domain-containing alpha polypeptide, SULFATE ION | | Authors: | Liu, L, Song, X, He, D, Komma, C, Kita, A, Verbasius, J.V, Bellamy, H, Miki, K, Czech, M.P, Zhou, G.W. | | Deposit date: | 2005-09-20 | | Release date: | 2005-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the C2 domain of class II phosphatidylinositide 3-kinase C2alpha.
J.Biol.Chem., 281, 2006
|
|
8EQM
 
 | | Structure of a dimeric photosystem II complex acclimated to far-red light | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Gisriel, C.J, Shen, G, Flesher, D.A, Kurashov, V, Golbeck, J.H, Brudvig, G.W, Amin, M, Bryant, D.A. | | Deposit date: | 2022-10-08 | | Release date: | 2022-12-28 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of a dimeric photosystem II complex from a cyanobacterium acclimated to far-red light.
J.Biol.Chem., 299, 2022
|
|
6PRZ
 
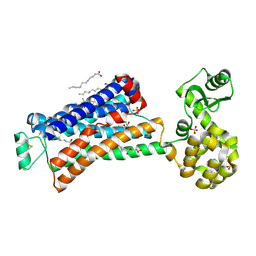 | | XFEL beta2 AR structure by ligand exchange from Alprenolol to Alprenolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
6PS1
 
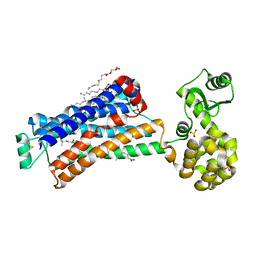 | | XFEL beta2 AR structure by ligand exchange from Alprenolol to Timolol. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-1-(tert-butylamino)-3-[(4-morpholin-4-yl-1,2,5-thiadiazol-3-yl)oxy]propan-2-ol, CHOLESTEROL, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
6B73
 
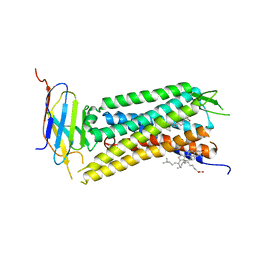 | | Crystal Structure of a nanobody-stabilized active state of the kappa-opioid receptor | | Descriptor: | CHOLESTEROL, N-[(5alpha,6beta)-17-(cyclopropylmethyl)-3-hydroxy-7,8-didehydro-4,5-epoxymorphinan-6-yl]-3-iodobenzamide, Nanobody, ... | | Authors: | Che, T, Majumdar, S, Zaidi, S.A, Kormos, C, McCorvy, J.D, Wang, S, Mosier, P.D, Uprety, R, Vardy, E, Krumm, B.E, Han, G.W, Lee, M.Y, Pardon, E, Steyaert, J, Huang, X.P, Strachan, R.T, Tribo, A.R, Pasternak, G.W, Carroll, I.F, Stevens, R.C, Cherezov, V, Katritch, V, Wacker, D, Roth, B.L. | | Deposit date: | 2017-10-03 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the Nanobody-Stabilized Active State of the Kappa Opioid Receptor.
Cell, 172, 2018
|
|
7S3D
 
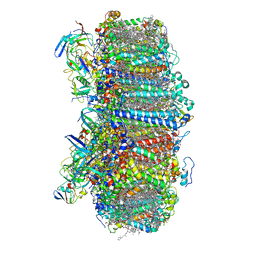 | | Structure of photosystem I with bound ferredoxin from Synechococcus sp. PCC 7335 acclimated to far-red light | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2Fe-2S ferredoxin-type domain-containing protein, ... | | Authors: | Gisriel, C.J, Flesher, D.A, Shen, G, Wang, J, Ho, M, Brudvig, G.W, Bryant, D.A. | | Deposit date: | 2021-09-05 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structure of a photosystem I-ferredoxin complex from a marine cyanobacterium provides insights into far-red light photoacclimation.
J.Biol.Chem., 298, 2021
|
|
7SA3
 
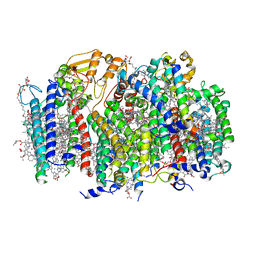 | | Structure of a monomeric photosystem II core complex from a cyanobacterium acclimated to far-red light | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Gisriel, C.J, Bryant, D.A, Brudvig, G.W. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Structure of a monomeric photosystem II core complex from a cyanobacterium acclimated to far-red light reveals the functions of chlorophylls d and f.
J.Biol.Chem., 298, 2021
|
|
7SK2
 
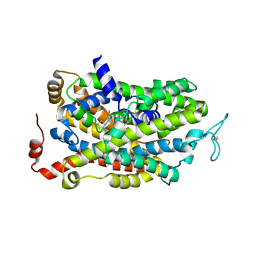 | | Human wildtype GABA reuptake transporter 1 in complex with tiagabine, inward-open conformation | | Descriptor: | Sodium- and chloride-dependent GABA transporter 1, Tiagabine | | Authors: | Gati, C, Motiwala, Z, Aduri, N.G, Shaye, H, Han, G.W, Cherezov, V. | | Deposit date: | 2021-10-19 | | Release date: | 2022-06-08 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis of GABA reuptake inhibition.
Nature, 606, 2022
|
|
2KDY
 
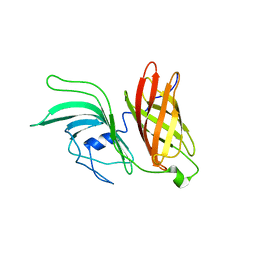 | | NMR structure of LP2086-B01 | | Descriptor: | Factor H binding protein variant B01_001 | | Authors: | Mascioni, A, Bentley, B.E, Camarda, R, Dilts, D.A, Fink, P, Gusarova, V, Hoiseth, S, Jacob, J, Lin, S.L, Malakian, K, McNeil, L.K, Mininni, T, Moy, F, Murphy, E, Novikova, E, Sigethy, S, Wen, Y, Zlotnick, G.W, Tsao, D.H.H. | | Deposit date: | 2009-01-20 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Immunogenic Properties of the Meningococcal Vaccine Candidate LP2086.
J.Biol.Chem., 284, 2009
|
|
2KLS
 
 | | Apo-form of the second Ca2+ binding domain of NCX1.4 | | Descriptor: | Sodium/calcium exchanger 1 | | Authors: | Hilge, M, Aelen, J, Foarce, A, Perrakis, A, Vuister, G.W. | | Deposit date: | 2009-07-08 | | Release date: | 2009-08-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Ca2+ regulation in the Na+/Ca2+ exchanger features a dual electrostatic switch mechanism.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KHR
 
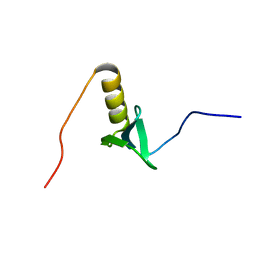 | |
