5MR4
 
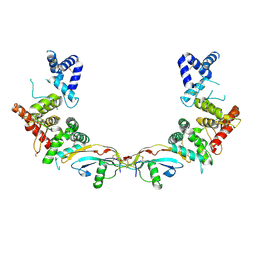 | | Ligand-receptor complex. | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, GDNF family receptor alpha-2, ... | | Authors: | Sandmark, J, Oster, L, Aagaard, A, Roth, R.G, Dahl, G. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and biophysical characterization of the human full-length neurturin-GFRa2 complex: A role for heparan sulfate in signaling.
J. Biol. Chem., 293, 2018
|
|
8QA7
 
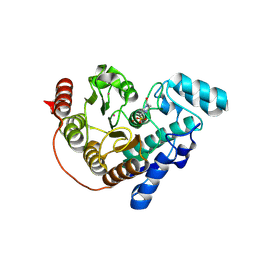 | |
5MR9
 
 | |
5MR5
 
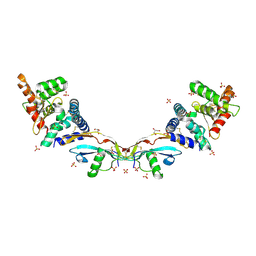 | | Ligand-receptor complex. | | Descriptor: | GDNF family receptor alpha-2, GLYCEROL, Neurturin, ... | | Authors: | Sandmark, J, Oster, L, Aagaard, A, Roth, R, Dahl, G. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biophysical characterization of the human full-length neurturin-GFRa2 complex: A role for heparan sulfate in signaling.
J. Biol. Chem., 293, 2018
|
|
1MLZ
 
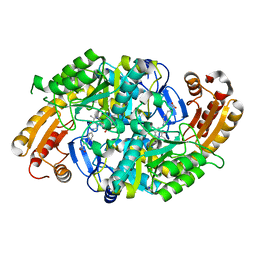 | | Crystal Structure of 7,8-Diaminopelargonic Acid Synthase in complex with the trans-isomer of amiclenomycin. | | Descriptor: | 7,8-diamino-pelargonic acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sandmark, J, Mann, S, Marquet, A, Schneider, G. | | Deposit date: | 2002-09-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the inhibition of the biosynthesis of biotin by the antibiotic amiclenomycin
J.Biol.Chem., 277, 2002
|
|
1MLY
 
 | | Crystal Structure of 7,8-Diaminopelargonic Acid Synthase in complex with the cis isomer of amiclenomycin | | Descriptor: | 7,8-diamino-pelargonic acid aminotransferase, CIS-AMICLENOMYCIN, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Sandmark, J, Mann, S, Marquet, A, Schneider, G. | | Deposit date: | 2002-09-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural basis for the inhibition of the biosynthesis of biotin by the antibiotic amiclenomycin
J.Biol.Chem., 277, 2002
|
|
1S08
 
 | | Crystal Structure of the D147N Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, SODIUM ION | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1S0A
 
 | | Crystal Structure of the Y17F Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, SODIUM ION | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1S09
 
 | | Crystal Structure of the Y144F Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, SODIUM ION | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1S06
 
 | | Crystal Structure of the R253K Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, SODIUM ION | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1S07
 
 | | Crystal Structure of the R253A Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, ISOPROPYL ALCOHOL, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
4BV5
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 4-(aminomethyl)-N-(benzenesulfonyl)cyclohexanecarboxamide, APOLIPOPROTEIN(A) | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4BVD
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 5-chloro-2-fluoro-N-[1-(4-piperidyl)pyrazol-4-yl]benzenesulfonamide, APOLIPOPROTEIN(A), CHLORIDE ION | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4BV7
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 3-(4-piperidyl)propanoic acid, ACETATE ION, APOLIPOPROTEIN(A) | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4BVC
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 3-(4-PIPERIDYL)-N-[2-(TRIFLUOROMETHOXY)PHENYL]SULFONYL-PROPANAMIDE, APOLIPOPROTEIN(A), CHLORIDE ION, ... | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4BVW
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 1,2,3,4-tetrahydroisoquinoline-6-carboxylic acid, 1,2-ETHANEDIOL, APOLIPOPROTEIN(A), ... | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4BVV
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, APOLIPOPROTEIN(A), SULFATE ION | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-28 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4UY1
 
 | | Novel pyrazole series of group X Secretory Phospholipase A2 (sPLA2-X) inhibitors | | Descriptor: | 5-(2,5-DIMETHYL-3-THIENYL)-1H-PYRAZOLE-3-CARBOXAMIDE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandmark, J, Oster, L, Hallberg, K, Bodin, C, Chen, H. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-15 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of a Novel Pyrazole Series of Group X Secreted Phospholipase A2 Inhibitor (Spla2X) Via Fragment Based Virtual Screening
Bioorg.Med.Chem.Lett., 24, 2014
|
|
8CJ7
 
 | | HDAC6 selective degraded (difluoromethyl)-1,3,4-oxadiazole substrate inhibitor | | Descriptor: | 6-[(5-pyridin-2-yl-1,2$l^{4},3,4-tetrazacyclopenta-1,3-dien-2-yl)methyl]pyridine-3-carbohydrazide, Histone deacetylase 6, IODIDE ION, ... | | Authors: | Sandmark, J, Ek, M, Ripa, L. | | Deposit date: | 2023-02-12 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Selective and Bioavailable HDAC6 2-(Difluoromethyl)-1,3,4-oxadiazole Substrate Inhibitors and Modeling of Their Bioactivation Mechanism.
J.Med.Chem., 66, 2023
|
|
4D7F
 
 | | Human FXIa in complex with small molecule inhibitors. | | Descriptor: | COAGULATION FACTOR XI, GLYCEROL, N-[(2S)-1-({4-[(diaminomethylidene)amino]butyl}amino)-1-oxo-3-phenylpropan-2-yl]-4-hydroxy-2-oxo-1,2-dihydroquinoline-6-carboxamide, ... | | Authors: | Sandmark, J, Oster, L, Jacso, T, Ullah, V, Redzick, A, Borjesson, U, Olsson, T, Norberg, M, Akerud, T. | | Deposit date: | 2014-11-24 | | Release date: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | From Mm Fragments to Nm Compounds Using Iloe-NMR to Guide Linking of Compounds in Fragment Based Drug Discovery (Fbdd).
To be Published
|
|
4D7G
 
 | | Human FXIa in complex with small molecule inhibitors. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COAGULATION FACTOR XIA, GLYCEROL, ... | | Authors: | Sandmark, J, Oster, L, Jacso, T, Ullah, V, Redzick, A, Borjesson, U, Olsson, T, Norberg, M, Akerud, T. | | Deposit date: | 2014-11-24 | | Release date: | 2016-01-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | From Mm Fragments to Nm Compounds Using Iloe-NMR to Guide Linking of Compounds in Fragment Based Drug Discovery (Fbdd).
To be Published
|
|
4CRF
 
 | | Creating novel F1 inhibitors through fragment based lead generation and structure aided drug design | | Descriptor: | CHLORIDE ION, Coagulation factor XIa light chain, GLYCEROL, ... | | Authors: | Sandmark, J, Oster, L, Fjellstrom, O, Akkaya, S, Beisel, H.G, Eriksson, P.O, Erixon, K, Gustafsson, D, Jurva, U, Kang, D, Karis, D, Knecht, W, Nerme, V, Nilsson, I, Olsson, T, Redzic, A, Roth, R, Tigerstrom, A. | | Deposit date: | 2014-02-26 | | Release date: | 2015-02-11 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Creating Novel Activated Factor Xi Inhibitors Through Fragment Based Lead Generation and Structure Aided Drug Design.
Plos One, 10, 2015
|
|
6G5J
 
 | | Secreted phospholipase A2 type X in complex with ligand | | Descriptor: | (3~{R})-3-[3-[2-aminocarbonyl-6-(trifluoromethyloxy)indol-1-yl]phenyl]butanoic acid, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandmark, J, Oster, L. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of Selective sPLA2-X Inhibitor (-)-2-{2-[Carbamoyl-6-(trifluoromethoxy)-1H-indol-1-yl]pyridine-2-yl}propanoic Acid.
ACS Med Chem Lett, 9, 2018
|
|
5G3N
 
 | | Discovery of a novel secreted phospholipase A2 (sPLA2) inhibitor. | | Descriptor: | 3-(5'-BENZYL-2'-CARBAMOYLBIPHENYL-3-YL)PROPANOIC ACID, CALCIUM ION, FORMIC ACID, ... | | Authors: | Sandmark, J, Bodin, C, Hallberg, K. | | Deposit date: | 2016-04-29 | | Release date: | 2016-09-14 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of Azd2716: A Novel Secreted Phospholipase A2 (Spla2) Inhibitor for the Treatment of Coronary Artery Disease
Acs Med.Chem.Lett., 7, 2016
|
|
5G3M
 
 | | Discovery of a novel secreted phospholipase A2 (sPLA2) inhibitor. | | Descriptor: | 4-BENZYLBENZAMIDE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sandmark, J, Bodin, C, Hallberg, K. | | Deposit date: | 2016-04-29 | | Release date: | 2016-09-14 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of Azd2716: A Novel Secreted Phospholipase A2 (Spla2) Inhibitor for the Treatment of Coronary Artery Disease
Acs Med.Chem.Lett., 7, 2016
|
|
