5LCY
 
 | | Formaldehyde-Responsive Regulator FrmR E64H variant from Salmonella enterica serovar Typhimurium | | Descriptor: | Frmr | | Authors: | Pohl, E, Robinson, N, Osman, D, Piergentili, C, Uson, I. | | Deposit date: | 2016-06-22 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Effectors and Sensory Sites of Formaldehyde-responsive Regulator FrmR and Metal-sensing Variant.
J.Biol.Chem., 291, 2016
|
|
2QZ8
 
 | | Crystal structure of Mycobacterium tuberculosis Leucine response regulatory protein (LrpA) | | Descriptor: | Probable transcriptional regulatory protein | | Authors: | Manchi, C.M.R, Gokulan, K, Ioerger, T, Jacobs Jr, W.R, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2007-08-16 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis LrpA, a leucine-responsive global regulator associated with starvation response.
Protein Sci., 17, 2008
|
|
2FMY
 
 | |
4E1N
 
 | |
4E1M
 
 | |
3C1B
 
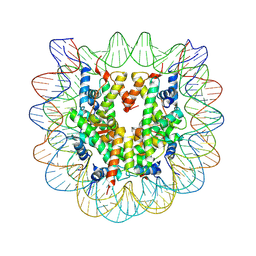 | | The effect of H3 K79 dimethylation and H4 K20 trimethylation on nucleosome and chromatin structure | | Descriptor: | Histone 2, H2bf, Histone H2A type 1, ... | | Authors: | Lu, X, Simon, M, Chodaparambil, J, Hansen, J, Shokat, K, Luger, K. | | Deposit date: | 2008-01-22 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure.
Nat.Struct.Mol.Biol., 15, 2008
|
|
8DEY
 
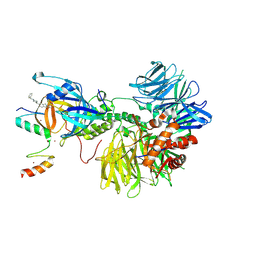 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2,3) and the molecular glue DKY709 | | Descriptor: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Ma, X, Ornelas, E, Clifton, M.C. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
9CAQ
 
 | |
6PB4
 
 | |
6D7R
 
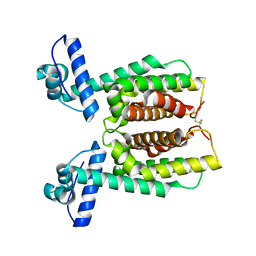 | | Crystal structure of an inactivate variant of the quorum-sensing master regulator HapR from protease-deficient non-O1, non-O139 Vibrio cholerae strain V2 | | Descriptor: | Hemagglutinin/protease regulatory protein | | Authors: | Cruite, J.T, Succo, P, Raychaudhuri, S, Kull, F.J. | | Deposit date: | 2018-04-25 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an inactive variant of the quorum-sensing master regulator HapR from the protease-deficient non-O1, non-O139 Vibrio cholerae strain V2.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4MCT
 
 | |
4MCX
 
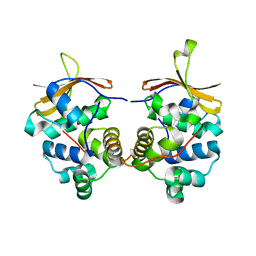 | |
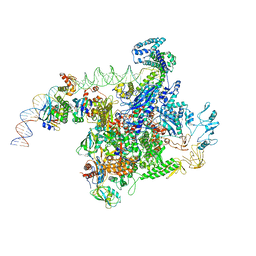
6PB6
 
 | |
4W97
 
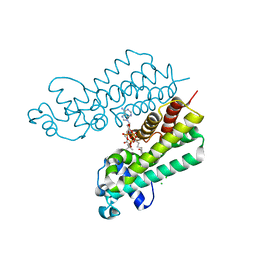 | | Structure of ketosteroid transcriptional regulator KstR2 of Mycobacterium tuberculosis | | Descriptor: | CHLORIDE ION, HTH-type transcriptional repressor KstR2, S-[2-[3-[[(2R)-4-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-4-oxidanyl-3-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3,3-dimethyl-2-oxidanyl-butanoyl]amino]propanoylamino]ethyl] 3-[(3aS,4S,7aS)-7a-methyl-1,5-bis(oxidanylidene)-2,3,3a,4,6,7-hexahydroinden-4-yl]propanethioate | | Authors: | Stogios, P.J, Evdokimova, E, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Characterization of a Ketosteroid Transcriptional Regulator of Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
7LBM
 
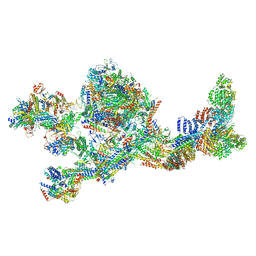 | | Structure of the human Mediator-bound transcription pre-initiation complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Abdella, R, Talyzina, A, He, Y. | | Deposit date: | 2021-01-08 | | Release date: | 2021-03-24 | | Last modified: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure of the human Mediator-bound transcription preinitiation complex.
Science, 372, 2021
|
|
1MM3
 
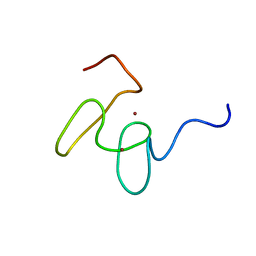 | | Solution structure of the 2nd PHD domain from Mi2b with C-terminal loop replaced by corresponding loop from WSTF | | Descriptor: | Mi2-beta(Chromodomain helicase-DNA-binding protein 4) and transcription factor WSTF, ZINC ION | | Authors: | Kwan, A.H.Y, Gell, D.A, Verger, A, Crossley, M, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2002-09-02 | | Release date: | 2003-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Engineering a Protein Scaffold from a PHD Finger
structure, 11, 2003
|
|
7ON1
 
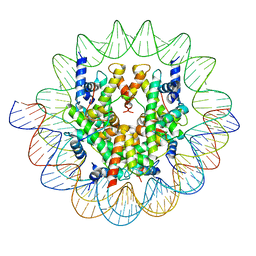 | | Cenp-A nucleosome in complex with Cenp-C | | Descriptor: | BJ4_G0006610.mRNA.1.CDS.1, BJ4_G0007000.mRNA.1.CDS.1, DNA (123-MER), ... | | Authors: | Yan, K, Yang, J, Zhang, Z, Barford, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-07-07 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cenp-A nucleosome in complex with Cenp-C
To Be Published
|
|
3T8R
 
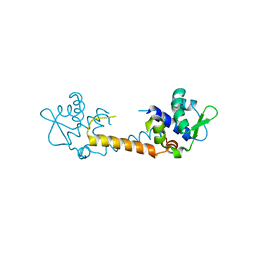 | | Crystal structure of Staphylococcus aureus CymR | | Descriptor: | Staphylococcus aureus CymR | | Authors: | He, C, Ji, Q. | | Deposit date: | 2011-08-01 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Staphylococcus aureus CymR Is a New Thiol-based Oxidation-sensing Regulator of Stress Resistance and Oxidative Response.
J.Biol.Chem., 287, 2012
|
|
3T22
 
 | |
6PB5
 
 | |
6L8F
 
 | |
6XII
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 24 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-06-20 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6ZTN
 
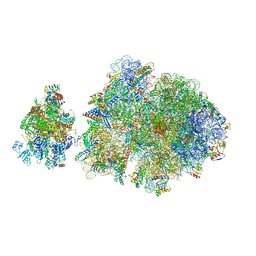 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (42 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
4MHQ
 
 | | Crystal structure of human primase catalytic subunit | | Descriptor: | CITRIC ACID, DNA primase small subunit, ZINC ION | | Authors: | Park, K.R, An, J.Y, Lee, Y, Youn, H.S, Lee, J.G, Kang, J.Y, Kim, T.G, Lim, J.J, Eom, S.H, Wang, J. | | Deposit date: | 2013-08-30 | | Release date: | 2014-09-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human primase catalytic subunit
To be Published
|
|
6XDR
 
 | | Escherichia coli transcription-translation complex B (TTC-B) containing an 27 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-06-11 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
