6W90
 
 | | De novo designed NTF2 fold protein NT-9 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, NTF2 fold protein loop-helix-loop design NT-9 | | Authors: | Thompson, M.C, Pan, X, Liu, L, Fraser, J.S, Kortemme, T. | | Deposit date: | 2020-03-21 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Expanding the space of protein geometries by computational design of de novo fold families.
Science, 369, 2020
|
|
5VLI
 
 | | Computationally designed inhibitor peptide HB1.6928.2.3 in complex with influenza hemagglutinin (A/PuertoRico/8/1934) | | Descriptor: | 2,5,8,11-TETRAOXATRIDECANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bernard, S.M, Wilson, I.A. | | Deposit date: | 2017-04-25 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Massively parallel de novo protein design for targeted therapeutics.
Nature, 550, 2017
|
|
1A48
 
 | | SAICAR SYNTHASE | | Descriptor: | PHOSPHORIBOSYLAMINOIMIDAZOLE-SUCCINOCARBOXAMIDE SYNTHASE, SULFATE ION | | Authors: | Levdikov, V.M, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K.S. | | Deposit date: | 1998-02-12 | | Release date: | 1999-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of SAICAR synthase: an enzyme in the de novo pathway of purine nucleotide biosynthesis.
Structure, 6, 1998
|
|
8PBO
 
 | | Deep interactome learning for generative drug design | | Descriptor: | 3-[2-fluoranyl-4-[3-[2-fluoranyl-4-(5-methyl-1,3,4-thiadiazol-2-yl)phenoxy]propoxy]phenyl]propanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Hakansson, M, Focht, D, Atz, K, Schneider, G. | | Deposit date: | 2023-06-09 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Prospective de novo drug design with deep interactome learning.
Nat Commun, 15, 2024
|
|
5VMR
 
 | |
5VID
 
 | |
6Q2F
 
 | | Structure of Rhamnosidase from Novosphingobium sp. PP1Y | | Descriptor: | Glycoside hydrolase family protein, SODIUM ION | | Authors: | Terry, B, Ha, J, Izzo, V, Sazinsky, M.H. | | Deposit date: | 2019-08-07 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.20000076 Å) | | Cite: | The crystal structure and insight into the substrate specificity of the alpha-L rhamnosidase RHA-P from Novosphingobium sp. PP1Y.
Arch.Biochem.Biophys., 679, 2019
|
|
1U0I
 
 | | IAAL-E3/K3 heterodimer | | Descriptor: | IAAL-E3, IAAL-K3 | | Authors: | Lindhout, D.A, Litowski, J.R, Mercier, P, Hodges, R.S, Sykes, B.D. | | Deposit date: | 2004-07-13 | | Release date: | 2004-10-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a highly stable de novo heterodimeric coiled-coil
Biopolymers, 75, 2004
|
|
8B16
 
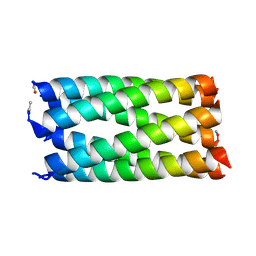 | |
8B15
 
 | |
5JD2
 
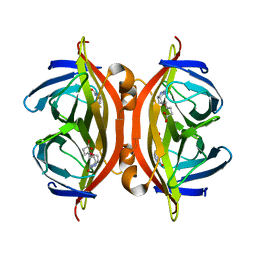 | | SFX structure of corestreptavidin-selenobiotin complex | | Descriptor: | 5-[(3aS,4S,6aR)-2-oxohexahydro-1H-selenopheno[3,4-d]imidazol-4-yl]pentanoic acid, Streptavidin | | Authors: | DeMirci, H, Hunter, M.S, Boutet, S. | | Deposit date: | 2016-04-15 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selenium single-wavelength anomalous diffraction de novo phasing using an X-ray-free electron laser.
Nat Commun, 7, 2016
|
|
5AYZ
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE PRODUCT NICOTINATE MONONUCLEOTIDE | | Descriptor: | NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
6L63
 
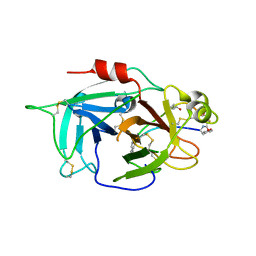 | | Human Coagulation Factor XIIa (FXIIa) bound with the macrocyclic peptide F3 containing two (1S,2S)-2-ACHC residues | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL GROUP, ... | | Authors: | Sengoku, T, Katoh, T, Hirata, K, Suga, H, Ogata, K. | | Deposit date: | 2019-10-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ribosomal synthesis and de novo discovery of bioactive foldamer peptides containing cyclic beta-amino acids.
Nat.Chem., 12, 2020
|
|
3O6Y
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | Descriptor: | Retro-Aldolase, SULFATE ION | | Authors: | Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J.K, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | Deposit date: | 2010-07-29 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
3NXF
 
 | | Robust computational design, optimization, and structural characterization of retroaldol enzymes | | Descriptor: | Retro-Aldolase, SULFATE ION | | Authors: | Althoff, E.A, Jiang, L, Wang, L, Lassila, J.K, Moody, J, Bolduc, J, Wang, Z.Z, Smith, M, Hari, S, Herschlag, D, Stoddard, B.L, Baker, D. | | Deposit date: | 2010-07-13 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
8E55
 
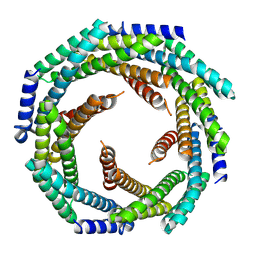 | |
2CW1
 
 | | Solution structure of the de novo-designed lambda Cro fold protein | | Descriptor: | SN4m | | Authors: | Isogai, Y, Ito, Y, Ikeya, T, Shiro, Y, Ota, M. | | Deposit date: | 2005-06-15 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Design of lambda Cro fold: solution structure of a monomeric variant of the de novo protein.
J.Mol.Biol., 354, 2005
|
|
4KVU
 
 | | Crystal structure of a 6-helix coiled coil CC-Hex-L17C-W224BF | | Descriptor: | 6-helix coiled coil CC-Hex-L17C-W224BF, GLYCEROL | | Authors: | Burton, A.J, Agnew, C, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2013-05-23 | | Release date: | 2013-08-21 | | Last modified: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Accessibility, Reactivity, and Selectivity of Side Chains within a Channel of de Novo Peptide Assembly.
J.Am.Chem.Soc., 135, 2013
|
|
4KVT
 
 | | Crystal structure of a 6-helix coiled coil CC-Hex-L24C | | Descriptor: | 6-helix coiled coil CC-Hex-L24C peptide | | Authors: | Burton, A.J, Agnew, C, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2013-05-23 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Accessibility, Reactivity, and Selectivity of Side Chains within a Channel of de Novo Peptide Assembly.
J.Am.Chem.Soc., 135, 2013
|
|
4KVV
 
 | | Crystal structure of an alkylated Cys mutant of CC-Hex | | Descriptor: | 6-HELIX COILED COIL CC-HEX-L24C PEPTIDE with an alkylated Cys mutation | | Authors: | Burton, A.J, Agnew, C, Brady, R.L, Woolfson, D.N. | | Deposit date: | 2013-05-23 | | Release date: | 2013-08-21 | | Last modified: | 2013-09-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Accessibility, Reactivity, and Selectivity of Side Chains within a Channel of de Novo Peptide Assembly.
J.Am.Chem.Soc., 135, 2013
|
|
8IJW
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-06 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8IJV
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-02 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[[5-chloranyl-2-(4-chlorophenyl)phenyl]methoxy]-N-methyl-but-2-yn-1-amine, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
8IJX
 
 | | Cryo-EM structure of the gastric proton pump with bound DQ-18 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[4-[(5-chloranyl-2-phenylmethoxy-phenyl)methoxy]phenyl]-N-methyl-methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Abe, K, Yokoshima, S, Yoshimori, A. | | Deposit date: | 2023-02-28 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.08 Å) | | Cite: | Deep learning driven de novo drug design based on gastric proton pump structures.
Commun Biol, 6, 2023
|
|
7RDH
 
 | |
8DQG
 
 | | Crystal structure of pyrrolysyl-tRNA synthetase from Methanomethylophilus alvus engineered for acridone amino acid (RS1) bound to AMPPNP and acridone | | Descriptor: | (2~{S})-2-azanyl-3-(9-oxidanylidene-10~{H}-acridin-2-yl)propanoic acid, AA_TRNA_LIGASE_II domain-containing protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gottfried-Lee, I, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2022-07-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity.
Acs Chem.Biol., 17, 2022
|
|
