6W6M
 
 | |
6PFZ
 
 | | Structure of a NAD-Dependent Persulfide Reductase from A. fulgidus | | Descriptor: | CALCIUM ION, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Sazinsky, M.H, Shabdar, S, Garcia-Constineiras, A, Crane III, E.J. | | Deposit date: | 2019-06-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.10003877 Å) | | Cite: | Structural and Kinetic Characterization of Hyperthermophilic NADH-Dependent Persulfide Reductase from Archaeoglobus fulgidus .
Archaea, 2021, 2021
|
|
2B8E
 
 | |
4F07
 
 | | Structure of the Styrene Monooxygenase Flavin Reductase (SMOB) from Pseudomonas putida S12 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NITRATE ION, SULFATE ION, ... | | Authors: | Sazinsky, M.H, Morrison, E, Kantz, A, Gassner, G. | | Deposit date: | 2012-05-03 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Mechanism of Styrene Monooxygenase Reductase: New Insight into the FAD-Transfer Reaction.
Biochemistry, 52, 2013
|
|
3N1X
 
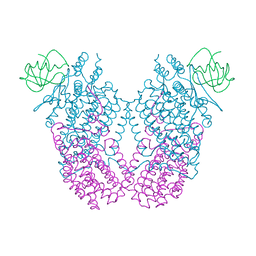 | | X-ray Crystal Structure of Toluene/o-Xylene Monooxygenase Hydroxylase T201C Mutant | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, HYDROXIDE ION, ... | | Authors: | Sazinsky, M.H, McCormick, M.S, Lippard, S.J. | | Deposit date: | 2010-05-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active Site Threonine Facilitates Proton Transfer during Dioxygen Activation at the Diiron Center of Toluene/o-Xylene Monooxygenase Hydroxylase.
J.Am.Chem.Soc., 132, 2010
|
|
2HC8
 
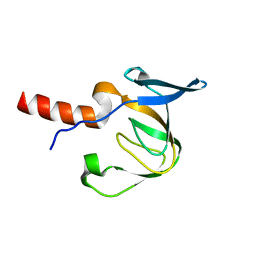 | | Structure of the A. fulgidus CopA A-domain | | Descriptor: | Cation-transporting ATPase, P-type | | Authors: | Sazinsky, M.H, Agawal, S, Arguello, J.M, Rosenzweig, A.C. | | Deposit date: | 2006-06-15 | | Release date: | 2006-09-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the Actuator Domain from the Archaeoglobus fulgidus Cu(+)-ATPase(,).
Biochemistry, 45, 2006
|
|
2HU9
 
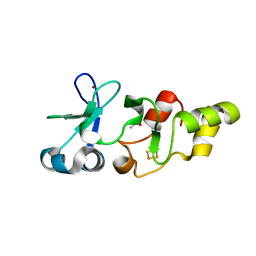 | | X-ray structure of the Archaeoglobus fulgidus CopZ N-terminal Domain | | Descriptor: | ACETIC ACID, FE2/S2 (INORGANIC) CLUSTER, Mercuric transport protein periplasmic component, ... | | Authors: | Sazinsky, M.H, LeMoine, B, Arguello, J.M, Rosenzweig, A.C. | | Deposit date: | 2006-07-26 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Characterization and structure of a Zn2+ and [2Fe-2S]-containing copper chaperone from Archaeoglobus fulgidus.
J.Biol.Chem., 282, 2007
|
|
2INN
 
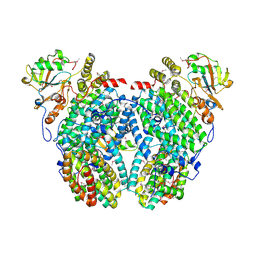 | | Structure of the Phenol Hydroxyalse-Regulatory Protein Complex | | Descriptor: | FE (III) ION, MOLYBDATE ION, Phenol hydroxylase component phL, ... | | Authors: | Sazinsky, M.H, Dunten, P.W, McCormick, M.S, Lippard, S.J. | | Deposit date: | 2006-10-08 | | Release date: | 2007-01-16 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray Structure of a Hydroxylase-Regulatory Protein Complex from a Hydrocarbon-Oxidizing Multicomponent Monooxygenase, Pseudomonas sp. OX1 Phenol Hydroxylase.
Biochemistry, 45, 2006
|
|
1T0R
 
 | | Crystal Structure of the Toluene/o-xylene Monooxygenase Hydroxuylase from Pseudomonas stutzeri-azide bound | | Descriptor: | AZIDE ION, FE (III) ION, HYDROXIDE ION, ... | | Authors: | Sazinsky, M.H, Bard, J, Di Donato, A, Lippard, S.J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase from Pseudomonas stutzeri OX1: INSIGHT INTO THE SUBSTRATE SPECIFICITY, SUBSTRATE CHANNELING, AND ACTIVE SITE TUNING OF MULTICOMPONENT MONOOXYGENASES.
J.Biol.Chem., 279, 2004
|
|
1T0S
 
 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase with 4-bromophenol bound | | Descriptor: | 4-BROMOPHENOL, FE (III) ION, HYDROXIDE ION, ... | | Authors: | Sazinsky, M.H, Bard, J, Di Donato, A, Lippard, S.J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase from Pseudomonas stutzeri OX1: INSIGHT INTO THE SUBSTRATE SPECIFICITY, SUBSTRATE CHANNELING, AND ACTIVE SITE TUNING OF MULTICOMPONENT MONOOXYGENASES.
J.Biol.Chem., 279, 2004
|
|
1T0Q
 
 | | Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase | | Descriptor: | FE (III) ION, HYDROXIDE ION, SULFANYLACETIC ACID, ... | | Authors: | Sazinsky, M.H, Bard, J, Di Donato, A, Lippard, S.J. | | Deposit date: | 2004-04-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Toluene/o-Xylene Monooxygenase Hydroxylase from Pseudomonas stutzeri OX1: INSIGHT INTO THE SUBSTRATE SPECIFICITY, SUBSTRATE CHANNELING, AND ACTIVE SITE TUNING OF MULTICOMPONENT MONOOXYGENASES.
J.Biol.Chem., 279, 2004
|
|
1XMF
 
 | | Structure of Mn(II)-Soaked Apo Methane Monooxygenase Hydroxylase Crystals from M. capsulatus (Bath) | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, Methane monooxygenase component A alpha chain, ... | | Authors: | Sazinsky, M.H, Merkx, M, Cadieux, E, Tang, S, Lippard, S.J. | | Deposit date: | 2004-10-02 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Preparation and X-ray Structures of Metal-Free, Dicobalt and Dimanganese Forms of Soluble Methane Monooxygenase Hydroxylase from Methylococcus capsulatus (Bath)
Biochemistry, 43, 2004
|
|
1XVG
 
 | |
1XMH
 
 | | Structure of Co(II) reconstituted methane monooxygenase hydroxylase from M. capsulatus (Bath) | | Descriptor: | COBALT (II) ION, Methane monooxygenase component A alpha chain, Methane monooxygenase component A beta chain, ... | | Authors: | Sazinsky, M.H, Merkx, M, Cadieux, E, Tang, S, Lippard, S.J. | | Deposit date: | 2004-10-02 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Preparation and X-ray Structures of Metal-Free, Dicobalt and Dimanganese Forms of Soluble Methane Monooxygenase Hydroxylase from Methylococcus capsulatus (Bath)
Biochemistry, 43, 2004
|
|
1XU5
 
 | | Soluble methane monooxygenase hydroxylase-phenol soaked | | Descriptor: | FE (III) ION, HYDROXIDE ION, Methane monooxygenase component A alpha chain, ... | | Authors: | Sazinsky, M.H, Lippard, S.J. | | Deposit date: | 2004-10-25 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Product Bound Structures of the Soluble Methane Monooxygenase Hydroxylase from Methylococcus capsulatus (Bath): Protein Motion in the Alpha-Subunit
J.Am.Chem.Soc., 127, 2005
|
|
1XVD
 
 | |
1XMG
 
 | | Crystal structure of apo methane monooxygenase hydroxylase from M. capsulatus (Bath) | | Descriptor: | CALCIUM ION, Methane monooxygenase component A alpha chain, Methane monooxygenase component A beta chain, ... | | Authors: | Sazinsky, M.H, Merkx, M, Cadieux, E, Tang, S, Lippard, S.J. | | Deposit date: | 2004-10-02 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Preparation and X-ray Structures of Metal-Free, Dicobalt and Dimanganese Forms of Soluble Methane Monooxygenase Hydroxylase from Methylococcus capsulatus (Bath)
Biochemistry, 43, 2004
|
|
1XVC
 
 | |
1XU3
 
 | | Soluble methane monooxygenase hydroxylase-soaked with bromophenol | | Descriptor: | 4-BROMOPHENOL, FE (III) ION, Methane monooxygenase component A alpha chain, ... | | Authors: | Sazinsky, M.H, Lippard, S.J. | | Deposit date: | 2004-10-25 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Product Bound Structures of the Soluble Methane Monooxygenase Hydroxylase from Methylococcus capsulatus (Bath): Protein Motion in the Alpha-Subunit
J.Am.Chem.Soc., 127, 2005
|
|
1XVB
 
 | |
1XVE
 
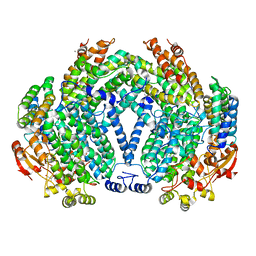 | |
1XVF
 
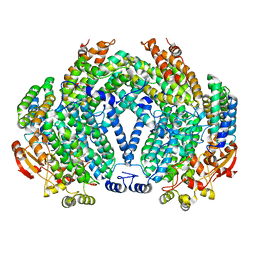 | |
3NTD
 
 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase C531S Mutant | | Descriptor: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | Authors: | Sazinsky, M.H, Warner, M.D, Lukose, V, Lee, K.H, Crane, E.J. | | Deposit date: | 2010-07-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
3NTA
 
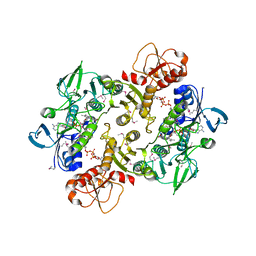 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase | | Descriptor: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | Authors: | Sazinsky, M.H, Crane, E.J, Warner, M.D, Lukose, V, Lee, K.H. | | Deposit date: | 2010-07-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
3NT6
 
 | | Structure of the Shewanella loihica PV-4 NADH-dependent persulfide reductase C43S/C531S Double Mutant | | Descriptor: | CHLORIDE ION, COENZYME A, FAD-dependent pyridine nucleotide-disulphide oxidoreductase, ... | | Authors: | Sazinsky, M.H, Crane, E.J, Warner, M.D, Lukose, V, Lee, K.H, Lopez, K. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of an NADH-Dependent Persulfide Reductase from Shewanella loihica PV-4: Implications for the Mechanism of Sulfur Respiration via FAD-Dependent Enzymes .
Biochemistry, 50, 2010
|
|
