1BBJ
 
 | |
1CID
 
 | |
2X8L
 
 | |
5MDH
 
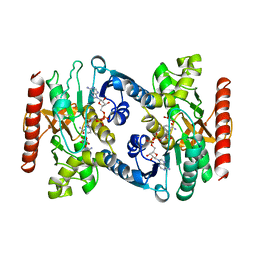 | | CRYSTAL STRUCTURE OF TERNARY COMPLEX OF PORCINE CYTOPLASMIC MALATE DEHYDROGENASE ALPHA-KETOMALONATE AND TNAD AT 2.4 ANGSTROMS RESOLUTION | | Descriptor: | ALPHA-KETOMALONIC ACID, MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Chapman, A.D.M, Cortes, A, Dafforn, T.R, Clarke, A.R, Brady, R.L. | | Deposit date: | 1998-10-08 | | Release date: | 1999-05-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of substrate specificity in malate dehydrogenases: crystal structure of a ternary complex of porcine cytoplasmic malate dehydrogenase, alpha-ketomalonate and tetrahydoNAD.
J.Mol.Biol., 285, 1999
|
|
1GSM
 
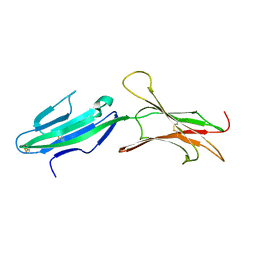 | | A reassessment of the MAdCAM-1 structure and its role in integrin recognition. | | Descriptor: | MUCOSAL ADDRESSIN CELL ADHESION MOLECULE-1 | | Authors: | Dando, J, Wilkinson, K.W, Ortlepp, S, King, D.J, Brady, R.L. | | Deposit date: | 2002-01-08 | | Release date: | 2002-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Reassessment of the Madcam-1 Structure and its Role in Integrin Recognition
Acta Crystallogr.,Sect.D, 58, 2002
|
|
4CUK
 
 | |
4CUJ
 
 | |
1WKO
 
 | |
1WKP
 
 | | Flowering locus t (ft) from arabidopsis thaliana | | Descriptor: | FLOWERING LOCUS T protein, SULFATE ION | | Authors: | Miller, D, Banfield, M.J, Winter, V.J, Brady, R.L. | | Deposit date: | 2004-06-01 | | Release date: | 2005-06-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A divergent external loop confers antagonistic activity on floral regulators FT and TFL1.
Embo J., 25, 2006
|
|
1XIV
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with 2-({4-chloro-[hydroxy(methoxy)methyl]cyclohexyl}amino)ethane-1,1,2-triol | | Descriptor: | 2-({4-CHLORO-2-[HYDROXY(METHOXY)METHYL]CYCLOHEXYL}AMINO)ETHANE-1,1,2-TRIOL, GLYCEROL, L-lactate dehydrogenase | | Authors: | Conners, R, Cameron, A, Read, J.A, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-09-22 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium
falciparum lactate dehydrogenase
MOL.BIOCHEM.PARASITOL., 142, 2005
|
|
4B7U
 
 | |
2PET
 
 | | Lutheran glycoprotein, N-terminal domains 1 and 2. | | Descriptor: | Lutheran blood group glycoprotein | | Authors: | Burton, N, Brady, R.L. | | Deposit date: | 2007-04-03 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Laminin 511/521-binding site on the Lutheran blood group glycoprotein is located at the flexible junction of Ig domains 2 and 3.
Blood, 110, 2007
|
|
2PF6
 
 | | Lutheran glycoprotein, N-terminal domains 1 and 2 | | Descriptor: | Lutheran blood group glycoprotein | | Authors: | Burton, N, Brady, R.L. | | Deposit date: | 2007-04-04 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Laminin 511/521-binding site on the Lutheran blood group glycoprotein is located at the flexible junction of Ig domains 2 and 3.
Blood, 110, 2007
|
|
2PLX
 
 | |
5KTV
 
 | |
5I4S
 
 | |
5LJ4
 
 | |
2CMY
 
 | | Crystal complex between bovine trypsin and Veronica hederifolia trypsin inhibitor | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, GLYCEROL, ... | | Authors: | Conners, R, Yardley, J.L, Konarev, A, Shewry, P, Brady, R.L. | | Deposit date: | 2006-05-16 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | An Unusual Helix-Turn-Helix Protease Inhibitory Motif in a Novel Trypsin Inhibitor from Seeds of Veronica (Veronica Hederifolia L.).
J.Biol.Chem., 282, 2007
|
|
2AYG
 
 | |
2AYE
 
 | |
2AYB
 
 | |
6YB2
 
 | |
6YB0
 
 | |
6YAZ
 
 | |
4ZQG
 
 | |
