2I7T
 
 | | Structure of human CPSF-73 | | Descriptor: | Cleavage and polyadenylation specificity factor 73 kDa subunit, SULFATE ION, ZINC ION | | Authors: | Mandel, C.R, Zhang, H, Tong, L. | | Deposit date: | 2006-08-31 | | Release date: | 2007-01-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Polyadenylation factor CPSF-73 is the pre-mRNA 3'-end-processing endonuclease.
Nature, 444, 2006
|
|
4O5A
 
 | | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140 | | Descriptor: | GLYCEROL, LacI family transcription regulator, SULFATE ION | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140.
To be Published
|
|
4KZJ
 
 | | Crystal Structure of TR3 LBD L449W Mutant | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F, Zhang, Q, Li, A, Tian, X, Cai, Q, Wang, W, Wang, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-05-30 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
4NB6
 
 | | Crystal structure of the ligand binding domain of RORC with T0901317 | | Descriptor: | N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, Nuclear receptor ROR-gamma | | Authors: | Hymowitz, S.G, Boenig-de Leon, G. | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based design of substituted hexafluoroisopropanol-arylsulfonamides as modulators of RORc.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4AH1
 
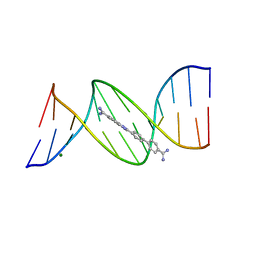 | | CRYSTAL STRUCTURE OF THE DB 921-D(CGCAAATTTGCG)2 COMPLEX AT 1.42 A RESOLUTION | | Descriptor: | 2-(4'-AMIDINOBIPHENYL-4-YL)-1H-BENZIMIDAZOLE-5-AMIDINE, 5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*GP)-3', MAGNESIUM ION | | Authors: | Munnur, D.G, Mitchell, E.P, Forsyth, V.T, Teixeira, S.C.M, Neidle, S. | | Deposit date: | 2012-02-02 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A Detailed Study of Water Network in the Minor Groove of D(Cgcgaattcgcg)2 and D(Cgcaaatttgcg)2 in Presence of Linear Groove Binders
To be Published
|
|
5HMC
 
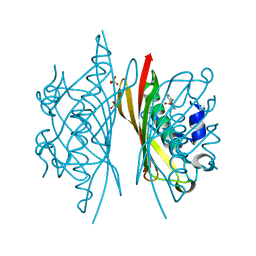 | | Crystal structure of S. sahachiroi AziG complexed with 5-methyl naphthoic acid | | Descriptor: | 5-methylnaphthalene-1-carboxylic acid, Azi13, SULFATE ION | | Authors: | Zhang, Y, Erb, M.S, Ealick, S.E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-02-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Polyketide Ring Expansion Mediated by a Thioesterase, Chain Elongation and Cyclization Domain, in Azinomycin Biosynthesis: Characterization of AziB and AziG.
Biochemistry, 55, 2016
|
|
4N8R
 
 | | Crystal structure of RXRa LBD complexed with a synthetic modulator K-8008 | | Descriptor: | 5-(2-{(1Z)-2-methyl-1-[4-(propan-2-yl)benzylidene]-1H-inden-3-yl}ethyl)-1H-tetrazole, Retinoic acid receptor RXR-alpha | | Authors: | Aleshin, A.E, Su, Y, Zhang, X, Liddington, R.C. | | Deposit date: | 2013-10-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Sulindac-Derived RXR alpha Modulators Inhibit Cancer Cell Growth by Binding to a Novel Site.
Chem.Biol., 21, 2014
|
|
4AGZ
 
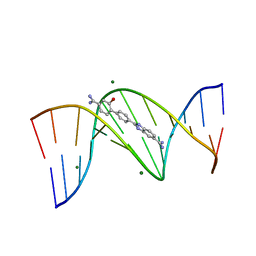 | | CRYSTAL STRUCTURE OF THE DB 985-D(CGCGAATTCGCG)2 COMPLEX AT 1.25 A RESOLUTION. | | Descriptor: | 2-(4'-CARBAMIMIDOYL-2'-HYDROXYBIPHENYL-4-YL)-1H-BENZIMIDAZOLE-5-CARBOXIMIDAMIDE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*GP)-3', MAGNESIUM ION | | Authors: | Munnur, D.G, Mitchell, E.P, Forsyth, V.T, Teixeira, S.C.M, Neidle, S. | | Deposit date: | 2012-02-02 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Detailed Study of Water Network in the Minor Groove of D(Cgcgaattcgcg)2 and D(Cgcaaatttgcg)2 in Presence of Linear Groove Binders
To be Published
|
|
4NR6
 
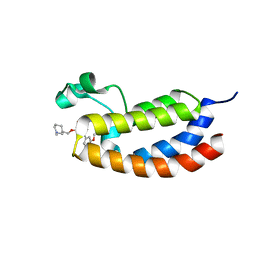 | | Crystal structure of the bromodomain of human CREBBP in complex with an oxazepin ligand | | Descriptor: | 1-[7-(3,4-dimethoxyphenyl)-9-{[(3R)-1-methylpiperidin-3-yl]methoxy}-2,3-dihydro-1,4-benzoxazepin-4(5H)-yl]propan-1-one, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Hay, D, Fedorov, O, Martin, S, Chaikuad, A, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of the bromodomain of human CREBBP in complex with an oxazepin ligand
To be Published
|
|
2YV0
 
 | | Structural and Thermodynamic Analyses of E. coli ribonuclease HI Variant with Quintuple Thermostabilizing Mutations | | Descriptor: | Ribonuclease HI | | Authors: | Haruki, M, Motegi, T, Tadokoro, T, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-04-06 | | Release date: | 2008-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and thermodynamic analyses of Escherichia coli RNase HI variant with quintuple thermostabilizing mutations.
Febs J., 274, 2007
|
|
2YCR
 
 | | Crystal structure of checkpoint kinase 2 in complex with inhibitor PV976 | | Descriptor: | 1,3-BIS{4-[(1E)-N-(4,5-DIHYDRO-1H-IMIDAZOL-2-YL)ETHANEHYDRAZONOYL]PHENYL}UREA, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C.R, Pommier, Y, Shoemaker, R.H, Zhang, G, Waugh, D.S. | | Deposit date: | 2011-03-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
4OZT
 
 | | crystal structure of the ligand binding domains of the Bovicola ovis ecdysone receptor EcR/USP heterodimer (PonA crystal) | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone receptor, N-ETHYLMALEIMIDE, ... | | Authors: | Ren, B, Peat, T.S, Streltsov, V.A, Pollard, M, Fernley, R, Grusovin, J, Seabrook, S, Pilling, P, Phan, T, Lu, L, Lovrecz, G.O, Graham, L.D, Hill, R.J. | | Deposit date: | 2014-02-19 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Unprecedented conformational flexibility revealed in the ligand-binding domains of the Bovicola ovis ecdysone receptor (EcR) and ultraspiracle (USP) subunits.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4O8F
 
 | | Crystal Structure of the complex between PPARgamma mutant R357A and rosiglitazone | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Chiaraluce, R, Consalvi, V, Lori, C, Loiodice, F, Laghezza, A, Pasquo, A, Cervoni, L, Aschi, M. | | Deposit date: | 2013-12-27 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the transactivation deficiency of the human PPAR gamma F360L mutant associated with familial partial lipodystrophy.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OIV
 
 | | Structural basis for small molecule NDB as a selective antagonist of FXR | | Descriptor: | Bile acid receptor, N-benzyl-N-(3-tert-butyl-4-hydroxyphenyl)-2,6-dichloro-4-(dimethylamino)benzamide | | Authors: | Xu, X, Chen, L, Hu, L, Shen, X. | | Deposit date: | 2014-01-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Small Molecule NDB (N-Benzyl-N-(3-(tert-butyl)-4-hydroxyphenyl)-2,6-dichloro-4-(dimethylamino) Benzamide) as a Selective Antagonist of Farnesoid X Receptor alpha (FXR alpha ) in Stabilizing the Homodimerization of the Receptor.
J.Biol.Chem., 290, 2015
|
|
4OJ4
 
 | |
4OC7
 
 | | Retinoic acid receptor alpha in complex with (E)-3-(3'-allyl-6-hydroxy-[1,1'-biphenyl]-3-yl)acrylic acid and a fragment of the coactivator TIF2 | | Descriptor: | (2E)-3-[6-hydroxy-3'-(prop-2-en-1-yl)biphenyl-3-yl]prop-2-enoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Leysen, S, Scheepstra, M, Brunsveld, L, Milroy, L.G, Ottmann, C. | | Deposit date: | 2014-01-08 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A natural-product switch for a dynamic protein interface.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2L16
 
 | |
3QB0
 
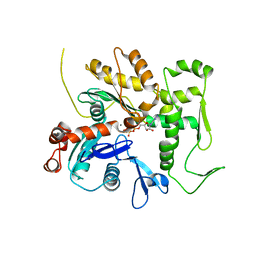 | | Crystal structure of Actin-related protein Arp4 from S. cerevisiae complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 4, CALCIUM ION | | Authors: | Fenn, S, Breitsprecher, D, Gerhold, C.B, Witte, G, Faix, J, Hopfner, K.P. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Structural biochemistry of nuclear actin-related proteins 4 and 8 reveals their interaction with actin.
Embo J., 30, 2011
|
|
8CBR
 
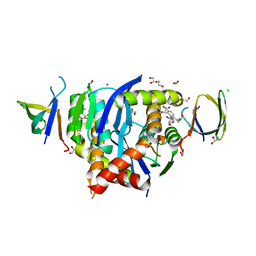 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor BDM-2 | | Descriptor: | (2S)-2-[3-cyclopropyl-2-(3,4-dihydro-2H-chromen-6-yl)-6-methyl-phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBU
 
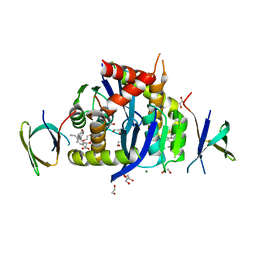 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT884 | | Descriptor: | (2S)-2-[3-cyclopropyl-6-methyl-2-(5-methyl-3,4-dihydro-2H-chromen-6-yl)phenyl]-2-cyclopropyloxy-ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBV
 
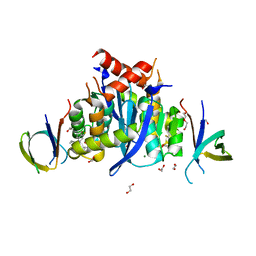 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT916 | | Descriptor: | (2~{S})-2-[3-cyclopropyl-2-(8-fluoranyl-5-methyl-3,4-dihydro-2~{H}-chromen-6-yl)-6-methyl-phenyl]-2-cyclopropyloxy-ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBT
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT872 | | Descriptor: | (2~{S})-2-[3-cyclopropyl-2-(3,4-dihydro-2~{H}-chromen-6-yl)-6-methyl-phenyl]-2-cyclopropyloxy-ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBS
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT871 | | Descriptor: | (2~{S})-2-[3-cyclopropyl-6-methyl-2-(5-methyl-3,4-dihydro-2~{H}-chromen-6-yl)phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
5Z0R
 
 | | Structural insight into the Zika virus capsid encapsulating the viral genome | | Descriptor: | Extracellular solute-binding protein family 1,viral genome protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Li, T, Zhao, Q, Yang, X, Chen, C, Yang, K, Wu, C, Zhang, T, Duan, Y, Xue, X, Mi, K, Ji, X, Wang, Z, Yang, H. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insight into the Zika virus capsid encapsulating the viral genome.
Cell Res., 28, 2018
|
|
2RQ5
 
 | |
