1OR3
 
 | |
2N5E
 
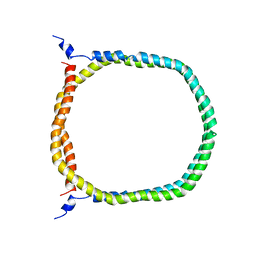 | | The 3D solution structure of discoidal high-density lipoprotein particles | | 分子名称: | Apolipoprotein A-I | | 著者 | Bibow, S, Polyhach, Y, Eichmann, C, Chi, C.N, Kowal, J, Stahlberg, H, Jeschke, G, Guentert, P, Riek, R. | | 登録日 | 2015-07-15 | | 公開日 | 2016-12-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of discoidal high-density lipoprotein particles with a shortened apolipoprotein A-I.
Nat.Struct.Mol.Biol., 24, 2017
|
|
8EQS
 
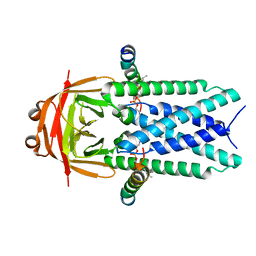 | | Structure of SARS-CoV-1 Orf3a in late endosome/lysosome-like environment, MSP1D1 nanodisc | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apolipoprotein A-I, ORF3a protein | | 著者 | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | 登録日 | 2022-10-09 | | 公開日 | 2023-02-08 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
7KJR
 
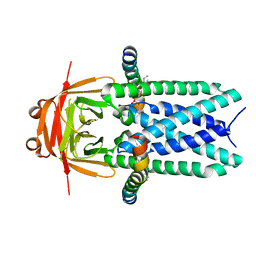 | | Cryo-EM structure of SARS-CoV-2 ORF3a | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Apolipoprotein A-I, ORF3a protein | | 著者 | Kern, D.M, Hoel, C.M, Kotecha, A, Brohawn, S.G. | | 登録日 | 2020-10-26 | | 公開日 | 2020-11-18 | | 最終更新日 | 2022-08-31 | | 実験手法 | ELECTRON MICROSCOPY (2.08 Å) | | 主引用文献 | Cryo-EM structure of SARS-CoV-2 ORF3a in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
8AX8
 
 | |
8AX9
 
 | |
8CE0
 
 | | N-terminal domain of human apolipoprotein E | | 分子名称: | GLYCEROL, Maltodextrin-binding protein,Apolipoprotein E | | 著者 | Marek, M, Nemergut, M. | | 登録日 | 2023-02-01 | | 公開日 | 2023-07-19 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Domino-like effect of C112R mutation on ApoE4 aggregation and its reduction by Alzheimer's Disease drug candidate.
Mol Neurodegener, 18, 2023
|
|
8CDY
 
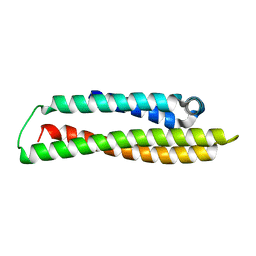 | | N-terminal domain of human apolipoprotein E | | 分子名称: | Maltodextrin-binding protein,Apolipoprotein E | | 著者 | Marek, M, Nemergut, M. | | 登録日 | 2023-02-01 | | 公開日 | 2023-07-19 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Domino-like effect of C112R mutation on ApoE4 aggregation and its reduction by Alzheimer's Disease drug candidate.
Mol Neurodegener, 18, 2023
|
|
4V6M
 
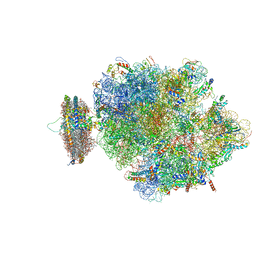 | | Structure of the ribosome-SecYE complex in the membrane environment | | 分子名称: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 16S RIBOSOMAL RNA, ... | | 著者 | Frauenfeld, J, Gumbart, J, van der Sluis, E.O, Funes, S, Gartmann, M, Beatrix, B, Mielke, T, Berninghausen, O, Becker, T, Schulten, K, Beckmann, R. | | 登録日 | 2011-02-08 | | 公開日 | 2014-07-09 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (7.1 Å) | | 主引用文献 | Cryo-EM structure of the ribosome-SecYE complex in the membrane environment.
Nat.Struct.Mol.Biol., 18, 2011
|
|
6W4F
 
 | | NMR-driven structure of KRAS4B-GDP homodimer on a lipid bilayer nanodisc | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | 著者 | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | 登録日 | 2020-03-10 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
8GRX
 
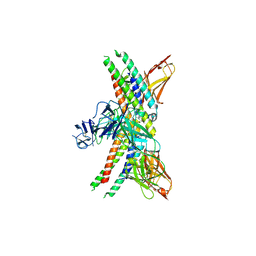 | | APOE4 receptor in complex with APOE4 NTD | | 分子名称: | Apolipoprotein E, Leukocyte immunoglobulin-like receptor subfamily A member 6 | | 著者 | Zhou, J, Wang, Y, Huang, G, Shi, Y. | | 登録日 | 2022-09-02 | | 公開日 | 2023-07-05 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | LilrB3 is a putative cell surface receptor of APOE4.
Cell Res., 33, 2023
|
|
6IWB
 
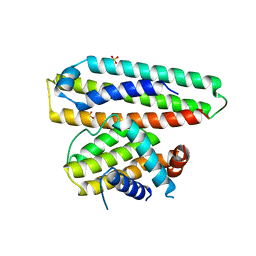 | | Crystal structure of a computationally designed protein (LD3) in complex with BCL-2 | | 分子名称: | Apolipoprotein E, Apoptosis regulator Bcl-2,Apoptosis regulator Bcl-2, SULFATE ION | | 著者 | Kim, S, Kwak, M.J, Oh, B.-H, Correia, B.E, Gainza, P. | | 登録日 | 2018-12-05 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | A computationally designed chimeric antigen receptor provides a small-molecule safety switch for T-cell therapy.
Nat.Biotechnol., 38, 2020
|
|
7RSC
 
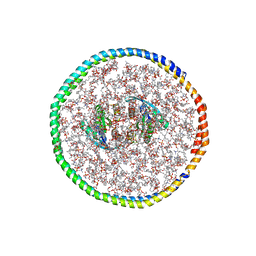 | | NMR-driven structure of the KRAS4B-G12D "alpha-alpha" dimer on a lipid bilayer nanodisc | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | 著者 | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | 登録日 | 2021-08-11 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
7RSE
 
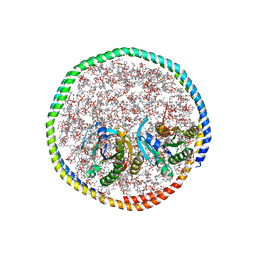 | | NMR-driven structure of the KRAS4B-G12D "alpha-beta" dimer on a lipid bilayer nanodisc | | 分子名称: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, GTPase KRas, ... | | 著者 | Lee, K, Enomoto, M, Gebregiworgis, T, Gasmi-Seabrook, G.M, Ikura, M, Marshall, C.B. | | 登録日 | 2021-08-11 | | 公開日 | 2021-09-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oncogenic KRAS G12D mutation promotes dimerization through a second, phosphatidylserine-dependent interface: a model for KRAS oligomerization.
Chem Sci, 12, 2021
|
|
6PTS
 
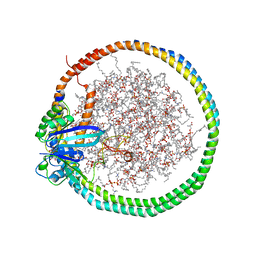 | | NMR data-driven model of KRas-GMPPNP:RBD-CRD complex tethered to a nanodisc (state A) | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | 著者 | Fang, Z, Lee, K, Gasmi-Seabrook, G, Ikura, M, Marshall, C.B. | | 登録日 | 2019-07-16 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PTW
 
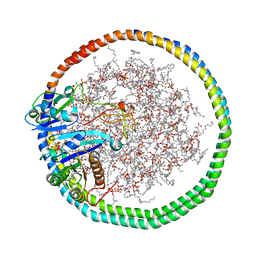 | | NMR data-driven model of KRas-GMPPNP:RBD-CRD complex tethered to a nanodisc (state B) | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | 著者 | Fang, Z, Lee, K, Gasmi-Seabrook, G, Ikura, M, Marshall, C.B. | | 登録日 | 2019-07-16 | | 公開日 | 2020-05-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6W4E
 
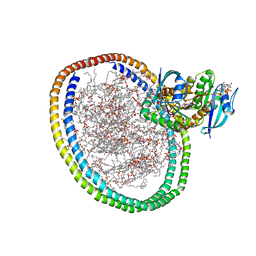 | | NMR-driven structure of KRAS4B-GTP homodimer on a lipid bilayer nanodisc | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Apolipoprotein A-I, ... | | 著者 | Lee, K, Fang, Z, Enomoto, M, Gasmi-Seabrook, G.M, Zheng, L, Marshall, C.B, Ikura, M. | | 登録日 | 2020-03-10 | | 公開日 | 2020-04-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Two Distinct Structures of Membrane-Associated Homodimers of GTP- and GDP-Bound KRAS4B Revealed by Paramagnetic Relaxation Enhancement.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6CC9
 
 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | 分子名称: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | 著者 | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | 登録日 | 2018-02-06 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6CCX
 
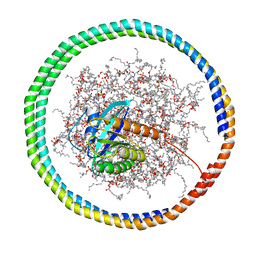 | | NMR data-driven model of GTPase KRas-GMPPNP:Cmpd2 complex tethered to a nanodisc | | 分子名称: | (2R,4S)-4-[(5-bromo-1H-indole-3-carbonyl)amino]-2-[(4-chlorophenyl)methyl]piperidin-1-ium, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, ... | | 著者 | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | 登録日 | 2018-02-07 | | 公開日 | 2018-09-05 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
6CM1
 
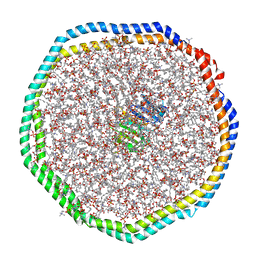 | |
6CLZ
 
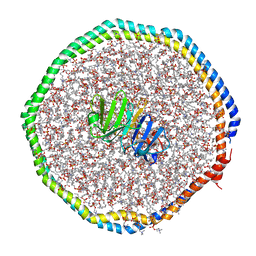 | |
6CCH
 
 | | NMR data-driven model of GTPase KRas-GMPPNP tethered to a nanodisc (E3 state) | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | 著者 | Fang, Z, Marshall, C.B, Nishikawa, T, Gossert, A.D, Jansen, J.M, Jahnke, W, Ikura, M. | | 登録日 | 2018-02-07 | | 公開日 | 2018-08-29 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Inhibition of K-RAS4B by a Unique Mechanism of Action: Stabilizing Membrane-Dependent Occlusion of the Effector-Binding Site.
Cell Chem Biol, 25, 2018
|
|
2MSD
 
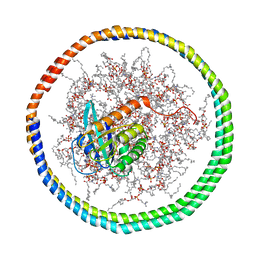 | | NMR data-driven model of GTPase KRas-GNP tethered to a lipid-bilayer nanodisc | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | 著者 | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | 登録日 | 2014-07-29 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSE
 
 | | NMR data-driven model of GTPase KRas-GNP:ARafRBD complex tethered to a lipid-bilayer nanodisc | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | 著者 | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | 登録日 | 2014-07-29 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2MSC
 
 | | NMR data-driven model of GTPase KRas-GDP tethered to a lipid-bilayer nanodisc | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Apolipoprotein A-I, GTPase KRas, ... | | 著者 | Mazhab-Jafari, M, Stathopoulos, P, Marshall, C, Ikura, M. | | 登録日 | 2014-07-29 | | 公開日 | 2015-06-03 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
