2MA9
 
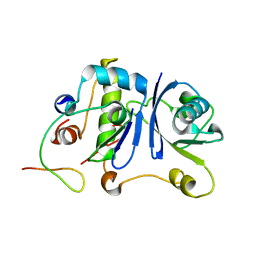 | | HIV-1 Vif SOCS-box and Elongin BC solution structure | | Descriptor: | Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, Virion infectivity factor | | Authors: | Lu, Z, Bergeron, J.R, Atkinson, R.A, Schaller, T, Veselkov, D.A, Oregioni, A, Yang, Y, Matthews, S.J, Malim, M.H, Sanderson, M.R. | | Deposit date: | 2013-07-01 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Insight into the HIV-1 Vif SOCS-box-ElonginBC interaction.
OPEN BIOLOGY, 3, 2013
|
|
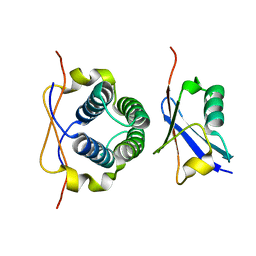
2LXC
 
 | |
7ZNT
 
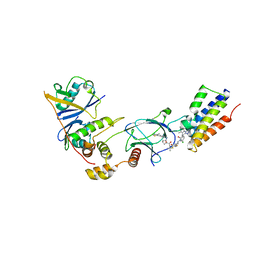 | | CRYSTAL STRUCTURE OF AT7 IN COMPLEX WITH THE SECOND BROMODOMAIN OF HUMAN BRD4 AND PVHL:ELONGINC:ELONGINB | | Descriptor: | (2~{S},4~{R})-1-[(2~{R})-3-[6-[2-[(9~{S})-7-(4-chlorophenyl)-4,5,13-trimethyl-3-thia-1,8,11,12-tetrazatricyclo[8.3.0.0^{2,6}]trideca-2(6),4,7,10,12-pentaen-9-yl]ethanoylamino]hexylsulfanyl]-2-[(1-fluoranylcyclopropyl)carbonylamino]-3-methyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Bromodomain-containing protein 4, Elongin-B, ... | | Authors: | Hughes, S.J, Casement, R, Ciulli, A. | | Deposit date: | 2022-04-22 | | Release date: | 2022-09-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Functional E3 ligase hotspots and resistance mechanisms to small-molecule degraders.
Nat.Chem.Biol., 19, 2023
|
|
7ZH4
 
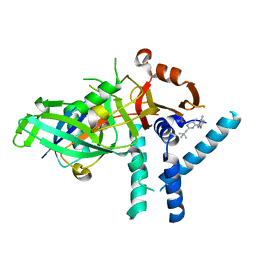 | | USP1 bound to ML323 and ubiquitin conjugated to FANCD2 (focused refinement) | | Descriptor: | 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phenyl]methyl]pyrimidin-4-amine, Ubiquitin carboxyl-terminal hydrolase 1, Ubiquitin-60S ribosomal protein L40, ... | | Authors: | Rennie, M.L, Walden, H. | | Deposit date: | 2022-04-05 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site.
Sci Adv, 8, 2022
|
|
7ZH3
 
 | |
7ZF1
 
 | | Structure of ubiquitinated FANCI in complex with FANCD2 and double-stranded DNA | | Descriptor: | DNA (61-MER), Fanconi anemia group D2 protein, Fanconi anemia group I protein, ... | | Authors: | Lemonidis, K, Rennie, M.L, Arkinson, C, Streetley, J, Clarke, M, Chaugule, V.K, Walden, H. | | Deposit date: | 2022-03-31 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structural and biochemical basis of interdependent FANCI-FANCD2 ubiquitination.
Embo J., 42, 2023
|
|
8JAL
 
 | | Structure of CRL2APPBP2 bound with RxxGP degron (dimer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAR
 
 | | Structure of CRL2APPBP2 bound with RxxGPAA degron (dimer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAS
 
 | | Structure of CRL2APPBP2 bound with RxxGPAA degron (tetramer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAQ
 
 | | Structure of CRL2APPBP2 bound with RxxGP degron (tetramer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-06 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAV
 
 | | Structure of CRL2APPBP2 bound with the C-degron of MRPL28 (tetramer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8JAU
 
 | | Structure of CRL2APPBP2 bound with the C-degron of MRPL28 (dimer) | | Descriptor: | Amyloid protein-binding protein 2, Cullin-2, Elongin-B, ... | | Authors: | Zhao, S, Zhang, K, Xu, C. | | Deposit date: | 2023-05-07 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Molecular basis for C-degron recognition by CRL2 APPBP2 ubiquitin ligase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8IVB
 
 | | K113-Ubiquitinated BAK | | Descriptor: | Bcl-2 homologous antagonist/killer, Ubiquitin | | Authors: | Dong, X, Cheng, P, Hou, Y.Z, Chen, Y.K, Liu, Z. | | Deposit date: | 2023-03-26 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Parkin-mediated ubiquitination inhibits BAK apoptotic activity by blocking its canonical hydrophobic groove.
Commun Biol, 6, 2023
|
|
7ZWL
 
 | | Crystal structure of human STING in complex with 3',3'-c-di-(2'F,2'dAMP) | | Descriptor: | 9-[(1~{R},6~{R},8~{R},9~{S},10~{R},15~{R},17~{R},18~{S})-17-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,11,13-tetraoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-8-yl]purin-6-amine, Stimulator of interferon protein, Ubiquitin-like protein SMT3 | | Authors: | Klima, M, Smola, M, Boura, E. | | Deposit date: | 2022-05-19 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human STING in complex with 3',3'-c-di-(2'F,2'dAMP)
To Be Published
|
|
8JE2
 
 | | Cryo-EM structure of neddylated Cul2-Rbx1-EloBC-FEM1B complexed with FNIP1-FLCN | | Descriptor: | Cullin-2, Elongin-B, Elongin-C, ... | | Authors: | Dai, Z, Liang, L, Yin, Y.X. | | Deposit date: | 2023-05-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
8JE1
 
 | | An asymmetry dimer of the Cul2-Rbx1-EloBC-FEM1B ubiquitin ligase complexed with BEX2 | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, Elongin-B, ... | | Authors: | Dai, Z, Liang, L, Yin, Y.X. | | Deposit date: | 2023-05-15 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural insights into the ubiquitylation strategy of the oligomeric CRL2 FEM1B E3 ubiquitin ligase.
Embo J., 43, 2024
|
|
8ITP
 
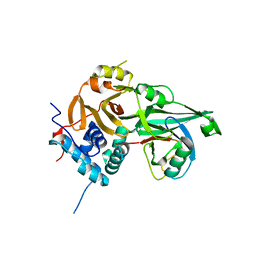 | |
8A9J
 
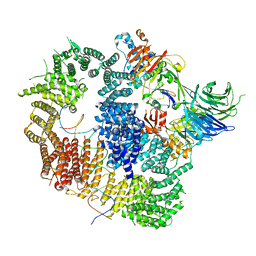 | |
8A9K
 
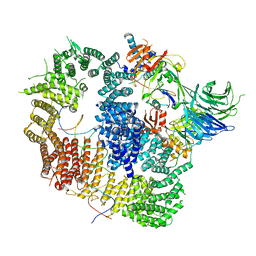 | | Cryo-EM structure of USP1-UAF1 bound to FANCI and mono-ubiquitinated FANCD2 with ML323 (consensus reconstruction) | | Descriptor: | 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phenyl]methyl]pyrimidin-4-amine, DNA (61-MER), Fanconi anemia group D2 protein, ... | | Authors: | Rennie, M.L, Walden, H. | | Deposit date: | 2022-06-28 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM reveals a mechanism of USP1 inhibition through a cryptic binding site.
Sci Adv, 8, 2022
|
|
8J1P
 
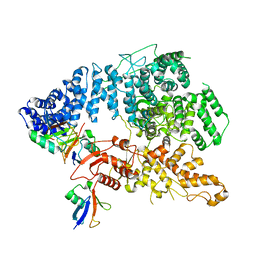 | | Cryo-EM structure of Ufd4 in complex with K29/48 triUb | | Descriptor: | Ubiquitin, Ubiquitin fusion degradation protein 4 | | Authors: | Ai, H.S, Mao, J.X, Wu, X.W, Pan, M, Liu, L. | | Deposit date: | 2023-04-13 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural Insights into the Molecular Mechanism of Ufd4-catalyzed Elongation of K48-linked Ubiquitin Chain through Lys29 Linkage
To Be Published
|
|
8IKV
 
 | |
8IKM
 
 | | Trans complex of phospho parkin | | Descriptor: | DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Lenka, D.R, Kumar, A. | | Deposit date: | 2023-02-28 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Additional feedforward mechanism of Parkin activation via binding of phospho-UBL and RING0 in trans.
Elife, 13, 2024
|
|
8IKT
 
 | | Ternary trans-complex of phospho-parkin with cis ACT and pUb | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-AMINOPROPANE, E3 ubiquitin-protein ligase parkin, ... | | Authors: | Lenka, D.R, Kumar, A. | | Deposit date: | 2023-03-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Additional feedforward mechanism of Parkin activation via binding of phospho-UBL and RING0 in trans.
Elife, 13, 2024
|
|
8BB3
 
 | | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with PEG linker (conformation #1) | | Descriptor: | 1,2-ETHANEDIOL, Elongin-B, Elongin-C, ... | | Authors: | Kraemer, A, Doelle, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with PEG linker (conformation #1)
To Be Published
|
|
8BB2
 
 | | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with PEG linker (conformation #2) | | Descriptor: | 1,2-ETHANEDIOL, Elongin-B, Elongin-C, ... | | Authors: | Kraemer, A, Doelle, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with PEG linker (conformation #2)
To Be Published
|
|
