5C51
 
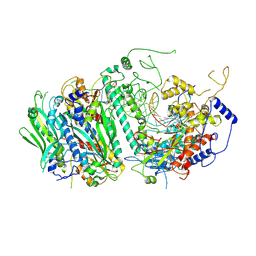 | | Probing the Structural and Molecular Basis of Nucleotide Selectivity by Human Mitochondrial DNA Polymerase gamma | | Descriptor: | DNA, DNA (5'-D(*(AD)P*AP*AP*AP*CP*GP*AP*GP*GP*GP*CP*CP*AP*GP*TP*GP*CP*CP*GP*TP*AP*C)-3'), DNA polymerase subunit gamma-1, ... | | Authors: | Sohl, C.D, Szymanski, M.R, Mislak, A.C, Shumate, C.K, Amiralaei, S, Schinazi, R.F, Anderson, K.S, Whitney, Y.Y. | | Deposit date: | 2015-06-19 | | Release date: | 2016-05-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.426 Å) | | Cite: | Probing the structural and molecular basis of nucleotide selectivity by human mitochondrial DNA polymerase gamma.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
104D
 
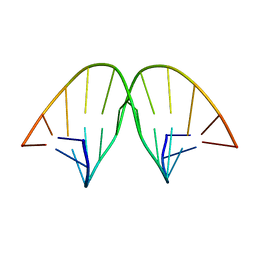 | |
1B79
 
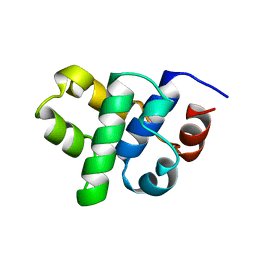 | |
6LWQ
 
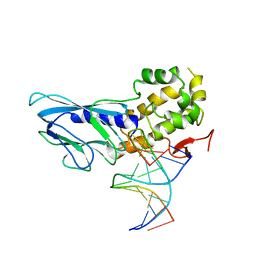 | | Crystal structure of human NEIL1(R242) bound to duplex DNA containing a C:T mismatch | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*TP*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*GP*G)-3'), Endonuclease 8-like 1 | | Authors: | Liu, M.H, Zhang, J, Zhu, C.X, Zhang, X.X, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2020-02-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | DNA repair glycosylase hNEIL1 triages damaged bases via competing interaction modes.
Nat Commun, 12, 2021
|
|
8EJ6
 
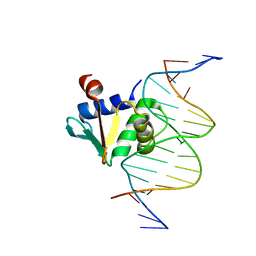 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAGAGGAATGGGG) | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*AP*GP*AP*GP*GP*AP*AP*TP*GP*GP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*CP*CP*AP*TP*TP*CP*CP*TP*CP*TP*TP*AP*T)-3'), Transcription factor PU.1 | | Authors: | Terrell, J.R, Poon, G.M.K. | | Deposit date: | 2022-09-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | DNA selection by the master transcription factor PU.1.
Cell Rep, 42, 2023
|
|
3KUY
 
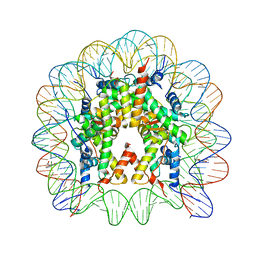 | |
5T4I
 
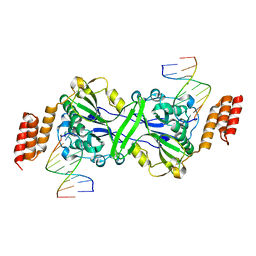 | |
5T5C
 
 | |
6CBR
 
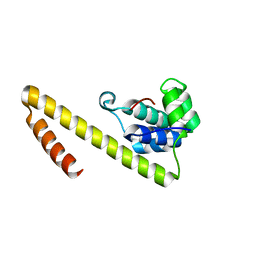 | |
8OJJ
 
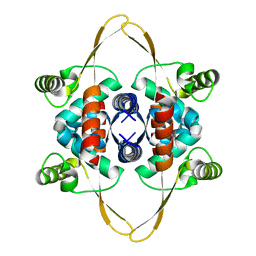 | | Cryo-EM structure of the DnaD-NTD tetramer | | Descriptor: | DNA replication protein DnaD | | Authors: | Winterhalter, C, Pelliciari, S, Cronin, N, Costa, T.R.D, Murray, H, Ilangovan, A. | | Deposit date: | 2023-03-24 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (5.47 Å) | | Cite: | The DNA replication initiation protein DnaD recognises a specific strand of the Bacillus subtilis chromosome origin.
Nucleic Acids Res., 51, 2023
|
|
4CRO
 
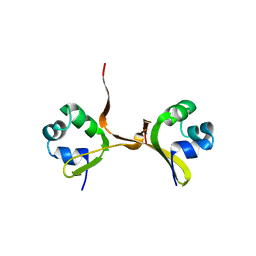 | | PROTEIN-DNA CONFORMATIONAL CHANGES IN THE CRYSTAL STRUCTURE OF A LAMBDA CRO-OPERATOR COMPLEX | | Descriptor: | DNA (5'-D(*TP*AP*TP*CP*AP*CP*CP*GP*CP*GP*GP*GP*TP*GP*AP*TP*A)-3'), PROTEIN (LAMBDA CRO) | | Authors: | Brennan, R.G, Roderick, S.L, Takeda, Y, Matthews, B.W. | | Deposit date: | 1992-01-15 | | Release date: | 1992-01-15 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Protein-DNA conformational changes in the crystal structure of a lambda Cro-operator complex.
Proc.Natl.Acad.Sci.USA, 87, 1990
|
|
1D17
 
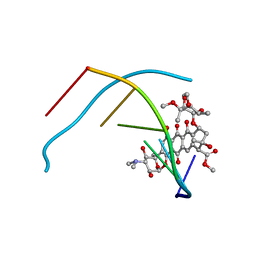 | | DNA-NOGALAMYCIN INTERACTIONS | | Descriptor: | DNA (5'-D(*(5CM)P*GP*TP*AP*(5CM)P*G)-3'), NOGALAMYCIN | | Authors: | Egli, M, Williams, L.D, Frederick, C.A, Rich, A. | | Deposit date: | 1990-08-08 | | Release date: | 1991-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA-nogalamycin interactions.
Biochemistry, 30, 1991
|
|
4NRV
 
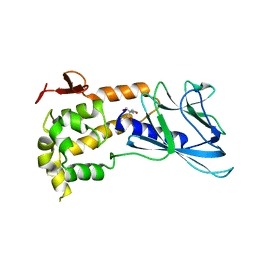 | | Crystal Structure of non-edited human NEIL1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endonuclease 8-like 1 | | Authors: | Prakash, A, Doublie, S. | | Deposit date: | 2013-11-27 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Genome and cancer single nucleotide polymorphisms of the human NEIL1 DNA glycosylase: Activity, structure, and the effect of editing.
Dna Repair, 14, 2014
|
|
3U6F
 
 | | Mouse TREX1 D200N mutant | | Descriptor: | 1,4-BUTANEDIOL, 5'-D(*GP*AP*CP*G)-3', MAGNESIUM ION, ... | | Authors: | Bailey, S.L, Harvey, S, Perrino, F.W, Hollis, T. | | Deposit date: | 2011-10-12 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Defects in DNA degradation revealed in crystal structures of TREX1 exonuclease mutations linked to autoimmune disease.
Dna Repair, 11, 2012
|
|
5GUJ
 
 | |
7M5O
 
 | | Cryo-EM structure of CasPhi-2 (Cas12j) bound to crRNA | | Descriptor: | CasPhi, ZINC ION, crRNA | | Authors: | Pausch, P, Soczek, K, Nogales, E, Doudna, J. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | DNA interference states of the hypercompact CRISPR-Cas Phi effector.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3U3Y
 
 | | Mouse TREX1 D200H mutant | | Descriptor: | 1,4-BUTANEDIOL, 5'-D(*GP*AP*CP*G)-3', CALCIUM ION, ... | | Authors: | Bailey, S.L, Harvey, S, Perrino, F.W, Hollis, T. | | Deposit date: | 2011-10-06 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Defects in DNA degradation revealed in crystal structures of TREX1 exonuclease mutations linked to autoimmune disease.
Dna Repair, 11, 2012
|
|
2DNJ
 
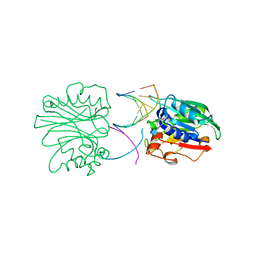 | |
6GDR
 
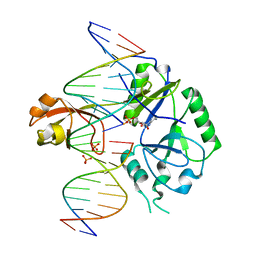 | | DNA binding with a minimal scaffold: Structure-function analysis of Lig E DNA ligases | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA, DNA (5'-D(*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*T)-3'), ... | | Authors: | Williamson, A, Grigic, M, Leiros, H.K.S. | | Deposit date: | 2018-04-24 | | Release date: | 2018-07-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | DNA binding with a minimal scaffold: structure-function analysis of Lig E DNA ligases.
Nucleic Acids Res., 46, 2018
|
|
1PAR
 
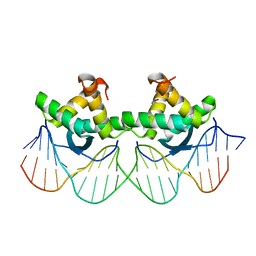 | | DNA RECOGNITION BY BETA-SHEETS IN THE ARC REPRESSOR-OPERATOR CRYSTAL STRUCTURE | | Descriptor: | DNA (5'-D(*AP*AP*TP*GP*AP*TP*AP*GP*AP*AP*GP*CP*AP*CP*TP*CP*T P*AP*CP*TP*AP*T)- 3'), DNA (5'-D(*TP*AP*TP*AP*GP*TP*AP*GP*AP*GP*TP*GP*CP*TP*TP*CP*T P*AP*TP*CP*AP*T)- 3'), PROTEIN (ARC REPRESSOR) | | Authors: | Raumann, B.E, Rould, M.A, Pabo, C.O, Sauer, R.T. | | Deposit date: | 1994-03-22 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DNA recognition by beta-sheets in the Arc repressor-operator crystal structure.
Nature, 367, 1994
|
|
5W35
 
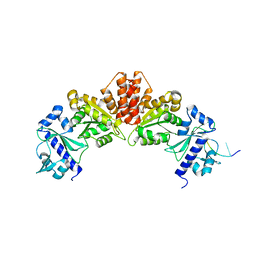 | |
5W34
 
 | |
5W33
 
 | |
5W36
 
 | |
6TEM
 
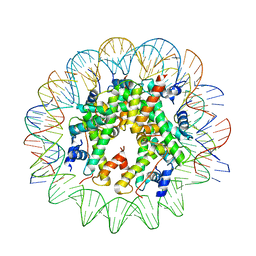 | | CENP-A nucleosome core particle with 145 base pairs of the Widom 601 sequence by cryo-EM | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3-like centromeric protein A, ... | | Authors: | Boopathi, R, Danev, R, Petosa, C, Bednar, J. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Phase-plate cryo-EM structure of the Widom 601 CENP-A nucleosome core particle reveals differential flexibility of the DNA ends.
Nucleic Acids Res., 48, 2020
|
|
