5BQB
 
 | | Crystal structure of Norrin, a Wnt signalling activator, Crystal Form III | | Descriptor: | CHLORIDE ION, CITRIC ACID, Norrin | | Authors: | Chang, T.-H, Hsieh, F.-L, Harlos, K, Jones, E.Y. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan.
Elife, 4, 2015
|
|
8CE7
 
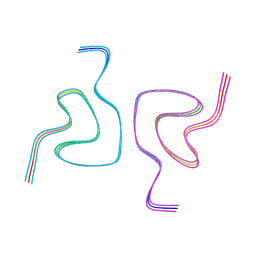 | | Type1 alpha-synuclein filament assembled in vitro by wild-type and mutant (7 residues insertion) protein | | Descriptor: | Alpha-synuclein | | Authors: | Yang, Y, Garringer, J.H, Shi, Y, Lovestam, S, Peak-Chew, S.Y, Zhang, X.J, Kotecha, A, Bacioglu, M, Koto, A, Takao, M, Spillantini, G.M, Ghetti, B, Vidal, R, Murzin, G.A, Scheres, H.W.S, Goedert, M. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-01 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | New SNCA mutation and structures of alpha-synuclein filaments from juvenile-onset synucleinopathy.
Acta Neuropathol, 145, 2023
|
|
8CEB
 
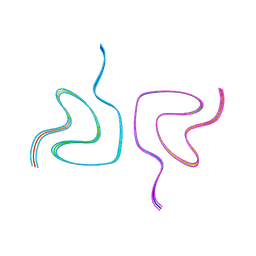 | | Type2 alpha-synuclein filament assembled in vitro by wild-type and mutant (7 residues insertion) protein | | Descriptor: | Alpha-synuclein | | Authors: | Yang, Y, Garringer, J.H, Shi, Y, Lovestam, S, Sew, P.C, Zhang, X.J, Kotecha, A, Bacioglu, M, Koto, A, Takao, M, Spillantini, G.M, Ghetti, B, Vidal, R, Murzin, G.A, Scheres, H.W.S, Goedert, M. | | Deposit date: | 2023-02-01 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | New SNCA mutation and structures of alpha-synuclein filaments from juvenile-onset synucleinopathy.
Acta Neuropathol, 145, 2023
|
|
5BRO
 
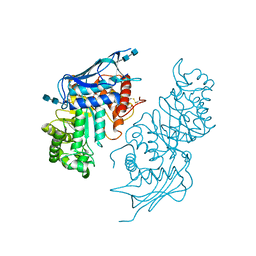 | | Crystal structure of modified HexB (modB) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosaminidase subunit beta, FORMIC ACID, ... | | Authors: | Kitakaze, K, Maita, N, Itoh, K. | | Deposit date: | 2015-06-01 | | Release date: | 2016-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Protease-resistant modified human beta-hexosaminidase B ameliorates symptoms in GM2 gangliosidosis model.
J.Clin.Invest., 126, 2016
|
|
5BQC
 
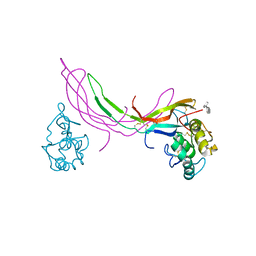 | | Crystal structure of Norrin in complex with the cysteine-rich domain of Frizzled 4 and sucrose octasulfate | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4, ... | | Authors: | Chang, T.-H, Hsieh, F.-L, Zebisch, M, Harlos, K, Jones, E.Y. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan.
Elife, 4, 2015
|
|
7TA8
 
 | | NMR structure of crosslinked cyclophilin A | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A | | Authors: | Lu, M, Toptygin, D, Xiang, Y, Shi, Y, Schwieters, C.D, Lipinski, E.C, Ahn, J, Byeon, I.-J.L, Gronenborn, A.M. | | Deposit date: | 2021-12-20 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The Magic of Linking Rings: Discovery of a Unique Photoinduced Fluorescent Protein Crosslink.
J.Am.Chem.Soc., 144, 2022
|
|
5NOW
 
 | | Structure of cyclophilin A in complex with pyridine-3,4-diamine | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, pyridine-3,4-diamine | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
1DI9
 
 | | THE STRUCTURE OF P38 MITOGEN-ACTIVATED PROTEIN KINASE IN COMPLEX WITH 4-[3-METHYLSULFANYLANILINO]-6,7-DIMETHOXYQUINAZOLINE | | Descriptor: | 4-[3-METHYLSULFANYLANILINO]-6,7-DIMETHOXYQUINAZOLINE, P38 KINASE | | Authors: | Shewchuk, L, Hassell, A, Kuyper, L.F. | | Deposit date: | 1999-11-29 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding mode of the 4-anilinoquinazoline class of protein kinase inhibitor: X-ray crystallographic studies of 4-anilinoquinazolines bound to cyclin-dependent kinase 2 and p38 kinase.
J.Med.Chem., 43, 2000
|
|
5CL1
 
 | | Complex structure of Norrin with human Frizzled 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-4, Maltose-binding periplasmic protein,Norrin | | Authors: | Wang, Z, Ke, J, Shen, G, Cheng, Z, Xu, H.E, Xu, W. | | Deposit date: | 2015-07-16 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of the Norrin-Frizzled 4 interaction.
Cell Res., 25, 2015
|
|
5NOS
 
 | | Structure of cyclophilin A in complex with 3-amino-1H-pyridin-2-one | | Descriptor: | 5-azanyl-3~{H}-pyridin-6-one, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
5NOQ
 
 | | Structure of cyclophilin A in complex with 3-chloropyridin-2-amine | | Descriptor: | 3-chloranylpyridin-2-amine, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
5NOU
 
 | | Structure of cyclophilin A in complex with hexahydropyrimidin-2-one | | Descriptor: | 1,3-diazinan-2-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
5NOR
 
 | | Structure of cyclophilin A in complex with 3-methylpyridin-2-amine | | Descriptor: | 3-methylpyridin-2-amine, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
5NOY
 
 | | Structure of cyclophilin A in complex with 3,4-diaminobenzamide | | Descriptor: | 3,4-bis(azanyl)benzamide, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
5NOT
 
 | | Structure of cyclophilin A in complex with 4-chloropyrimidin-5-amine | | Descriptor: | 4-chloranylpyrimidin-5-amine, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
5NOX
 
 | | Structure of cyclophilin A in complex with 2-chloropyridin-3-amine | | Descriptor: | 2-chloranylpyridin-3-amine, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
6SFI
 
 | | Crystal structure of p38 alpha in complex with compound 75 (MCP33) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]-2-[[[1-(2-methylphenyl)pyrazol-4-yl]carbonylamino]methyl]-1,3-thiazole-5-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
8F86
 
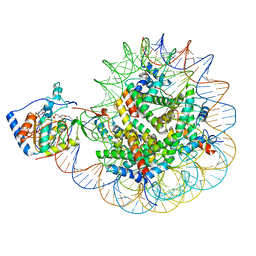 | | SIRT6 bound to an H3K9Ac nucleosome | | Descriptor: | DNA (148-MER), Histone H2A type 1, Histone H2B, ... | | Authors: | Markert, J, Whedon, S, Wang, Z, Cole, P, Farnung, L. | | Deposit date: | 2022-11-21 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Sirtuin 6-Catalyzed Nucleosome Deacetylation.
J.Am.Chem.Soc., 145, 2023
|
|
6SFJ
 
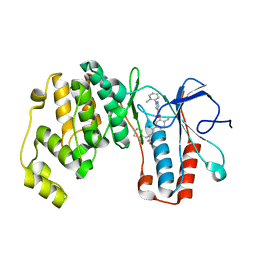 | | Crystal structure of p38 alpha in complex with compound 77 (MCP41) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-(2-methylphenyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
7P27
 
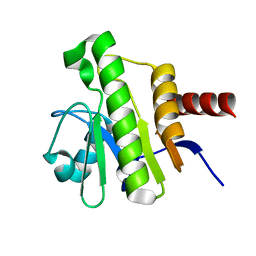 | | NMR solution structure of Chikungunya virus macro domain | | Descriptor: | Polyprotein P1234 | | Authors: | Lykouras, M.V, Tsika, A.C, Papageorgiou, N, Canard, B, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2021-07-04 | | Release date: | 2022-07-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Binding Adaptation of GS-441524 Diversifies Macro Domains and Downregulates SARS-CoV-2 de-MARylation Capacity.
J.Mol.Biol., 434, 2022
|
|
6SFO
 
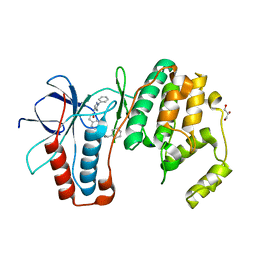 | | MAPK14 with bound inhibitor SR-318 | | Descriptor: | 5-azanyl-~{N}-[[4-(3-cyclohexylpropylcarbamoyl)phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, GLYCEROL, Mitogen-activated protein kinase 14, ... | | Authors: | Schroeder, M, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | MAPK14 with bound inhibitor SR-318
To Be Published
|
|
5NOZ
 
 | | Structure of cyclophilin A in complex with 3,4-diaminobenzohydrazide | | Descriptor: | 3,4-bis(azanyl)benzohydrazide, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Georgiou, C, Mcnae, I.W, Ioannidis, H, Julien, M, Walkinshaw, M.D. | | Deposit date: | 2017-04-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Pushing the Limits of Detection of Weak Binding Using Fragment-Based Drug Discovery: Identification of New Cyclophilin Binders.
J. Mol. Biol., 429, 2017
|
|
6SFK
 
 | | Crystal structure of p38 alpha in complex with compound 81 (MCP42) | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-phenyl-5-(trifluoromethyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
5I42
 
 | | Structure of HIV-1 Reverse Transcriptase in complex with a DNA aptamer, AZTTP, and CA(2+) ion | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (38-MER), ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2016-02-11 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Conformational States of HIV-1 Reverse Transcriptase for Nucleotide Incorporation vs Pyrophosphorolysis-Binding of Foscarnet.
Acs Chem.Biol., 11, 2016
|
|
7R1D
 
 | | Structure of MuvB complex | | Descriptor: | Histone-binding protein RBBP4, Protein lin-37 homolog, Protein lin-9 homolog | | Authors: | Koliopoulos, M.G, Alfieri, C. | | Deposit date: | 2022-02-02 | | Release date: | 2022-08-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a nucleosome-bound MuvB transcription factor complex reveals DNA remodelling.
Nat Commun, 13, 2022
|
|
