3FIW
 
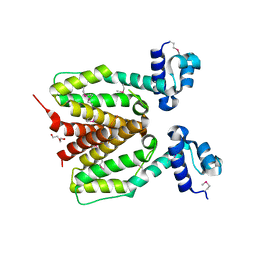 | | Structure of SCO0253, a Tetr-family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | DI(HYDROXYETHYL)ETHER, Putative tetR-family transcriptional regulator | | Authors: | Singer, A.U, Xu, X, Chang, C, Gu, J, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-12 | | Release date: | 2009-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SCO0253, a Tetr-family transcriptional regulator from Streptomyces coelicolor
To be Published
|
|
7UYF
 
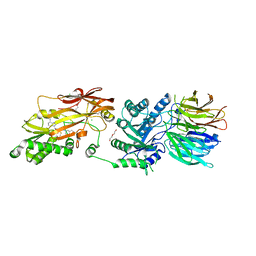 | | Human PRMT5:MEP50 structure with Fragment 4 and MTA Bound | | Descriptor: | 4-methyl-1,5-naphthyridin-2-amine, 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, ... | | Authors: | Gunn, R.J, Lawson, J.D, Smith, C.R. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
7UY1
 
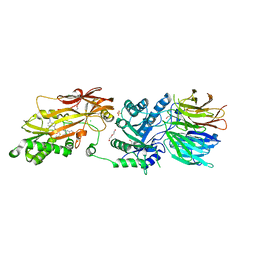 | | HUMAN PRMT5:MEP50 COMPLEX WITH MTA and Fragment 5 Bound | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-1,5-naphthyridin-2-amine, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ... | | Authors: | Gunn, R.J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
6RLQ
 
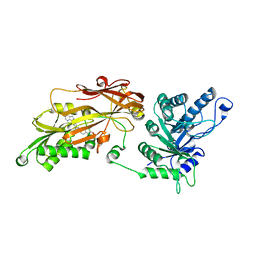 | | CRYSTAL STRUCTURE OF THE HUMAN PRMT5:MEP50 COMPLEX with JNJ45031882 | | Descriptor: | (1~{S},2~{R},3~{S},5~{R})-3-[2-(2-azanyl-3-bromanyl-quinolin-7-yl)ethyl]-5-(4-azanylpyrrolo[2,3-d]pyrimidin-7-yl)cyclop entane-1,2-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | Brown, D.G, Robinson, C.M, Pande, V. | | Deposit date: | 2019-05-02 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | CRYSTAL STRUCTURE OF THE HUMAN PRMT5:MEP50 COMPLEX with JNJ45031882
To Be Published
|
|
5ME0
 
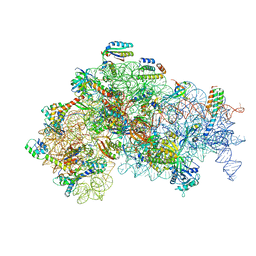 | | Structure of the 30S Pre-Initiation Complex 1 (30S IC-1) Stalled by GE81112 | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Fabbretti, A, Kaminishi, T, Iturrioz, I, Brandi, L, Gil Carton, D, Gualerzi, C, Fucini, P, Connell, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Structure of a 30S pre-initiation complex stalled by GE81112 reveals structural parallels in bacterial and eukaryotic protein synthesis initiation pathways.
Nucleic Acids Res., 45, 2017
|
|
6RLL
 
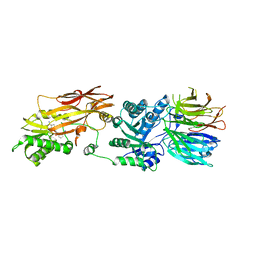 | | CRYSTAL STRUCTURE OF THE HUMAN PRMT5:MEP50 COMPLEX with JNJ44064146 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(4-azanylpyrrolo[2,3-d]pyrimidin-7-yl)-5-(1,8-diazaspiro[4.5]decan-1-ylmethyl)oxolane-3,4-diol, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | Brown, D.G, Robinson, C.M, Pande, V. | | Deposit date: | 2019-05-02 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A chemical probe for the methyl transferase PRMT5 with a novel
binding mode
To Be Published
|
|
8FX4
 
 | | GC-C-Hsp90-Cdc37 regulatory complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Guanylyl cyclase C, Heat shock protein HSP 90-beta, ... | | Authors: | Caveney, N.A, Garcia, K.C. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-12 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into guanylyl cyclase receptor hijacking of the kinase-Hsp90 regulatory mechanism.
Elife, 12, 2023
|
|
5TGM
 
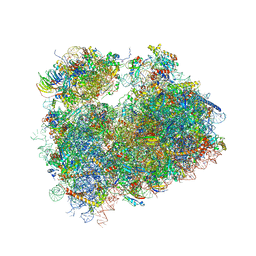 | | Crystal structure of the S.cerevisiae 80S ribosome in complex with the A-site bound aminoacyl-tRNA analog ACCA-Pro | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3'-amino-3'-deoxyadenosine 5'-(dihydrogen phosphate), ... | | Authors: | Melnikov, S, Mailliot, J, Yusupov, M. | | Deposit date: | 2016-09-28 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular insights into protein synthesis with proline residues.
EMBO Rep., 17, 2016
|
|
8G1U
 
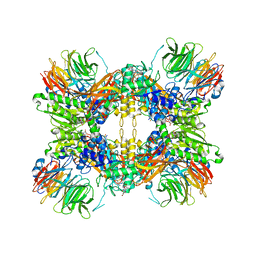 | | Structure of the methylosome-Lsm10/11 complex | | Descriptor: | ADENOSINE, Methylosome protein 50, Methylosome subunit pICln, ... | | Authors: | Lin, M, Paige, A, Tong, L. | | Deposit date: | 2023-02-03 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | In vitro methylation of the U7 snRNP subunits Lsm11 and SmE by the PRMT5/MEP50/pICln methylosome.
Rna, 29, 2023
|
|
5ME1
 
 | | Structure of the 30S Pre-Initiation Complex 2 (30S IC-2) Stalled by GE81112 | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Lopez-Alonso, J.P, Fabbretti, A, Kaminishi, T, Iturrioz, I, Brandi, L, Gil Carton, D, Gualerzi, C, Fucini, P, Connell, S. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-11 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Structure of a 30S pre-initiation complex stalled by GE81112 reveals structural parallels in bacterial and eukaryotic protein synthesis initiation pathways.
Nucleic Acids Res., 45, 2017
|
|
7Q0P
 
 | | Structure of the Candida albicans 80S ribosome in complex with anisomycin | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-15 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7Q0F
 
 | | Structure of Candida albicans 80S ribosome in complex with phyllanthoside | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7Q08
 
 | | Structure of Candida albicans 80S ribosome in complex with cycloheximide | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7PZY
 
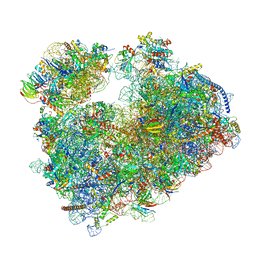 | | Structure of the vacant Candida albicans 80S ribosome | | Descriptor: | 1,4-DIAMINOBUTANE, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
7Q0R
 
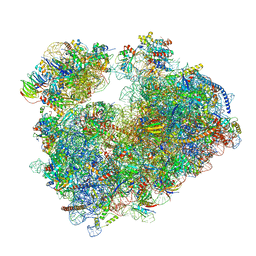 | | Structure of the Candida albicans 80S ribosome in complex with blasticidin s | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-16 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
8Q7N
 
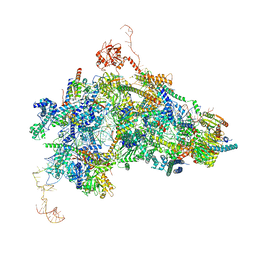 | | cryo-EM structure of the human spliceosomal B complex protomer (tri-snRNP core region) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, MINX pre-mRNA, Microfibrillar-associated protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2023-08-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM analyses of dimerized spliceosomes provide new insights into the functions of B complex proteins.
Embo J., 43, 2024
|
|
3DOX
 
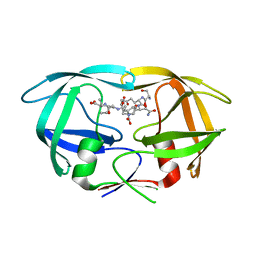 | | X-ray structure of HIV-1 protease in situ product complex | | Descriptor: | A PEPTIDE SUBSTRATE-PIV, A PEPTIDE SUBSTRATE-SQNY, HIV-1 PROTEASE | | Authors: | Hosur, M.V, Ferrer, J.-L, Das, A, Prashar, V, Bihani, S. | | Deposit date: | 2008-07-07 | | Release date: | 2008-09-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of HIV-1 protease in situ product complex
Proteins, 74, 2009
|
|
3BHT
 
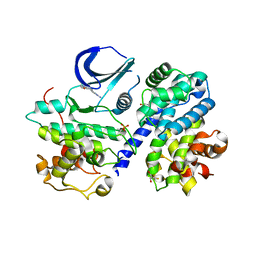 | | Structure of phosphorylated Thr160 CDK2/cyclin A in complex with the inhibitor meriolin 3 | | Descriptor: | 4-(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-2-amine, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Echalier, A, Bettayeb, K, Ferandin, Y, Lozach, O, Clement, M, Valette, A, Liger, F, Marquet, B, Morris, J.C, Endicott, J.A, Joseph, B, Meijer, L. | | Deposit date: | 2007-11-29 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Meriolins, a new class of cell death inducing kinase inhibitors with enhanced selectivity for cyclin-dependent kinases
Cancer Res., 67, 2007
|
|
7U30
 
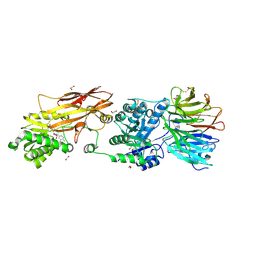 | | PRMT5:MEP50 Complexed with Cyclonucleoside Compound 1 | | Descriptor: | (9R,10R,11S,12R,13R,14R)-4-amino-9-(3,4-difluorophenyl)-6,7,8,9,10,11,12,13-octahydro-10,13-epoxy[1,3]diazecino[1,2-e]purine-11,12-diol, 1,2-ETHANEDIOL, Methylosome protein 50, ... | | Authors: | Palte, R.L. | | Deposit date: | 2022-02-25 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design and synthesis of unprecedented 9- and 10-membered cyclonucleosides with PRMT5 inhibitory activity.
Bioorg.Med.Chem., 66, 2022
|
|
3BHV
 
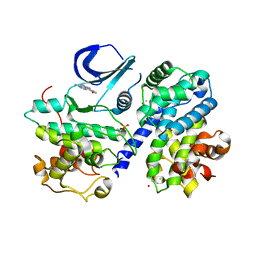 | | Structure of phosphorylated Thr160 CDK2/cyclin A in complex with the inhibitor variolin B | | Descriptor: | 9-amino-5-(2-aminopyrimidin-4-yl)pyrido[3',2':4,5]pyrrolo[1,2-c]pyrimidin-4-ol, Cell division protein kinase 2, Cyclin-A2, ... | | Authors: | Echalier, A, Bettayeb, K, Ferandin, Y, Lozach, O, Clement, M, Valette, A, Liger, F, Marquet, B, Morris, J.C, Endicott, J.A, Joseph, B, Meijer, L. | | Deposit date: | 2007-11-29 | | Release date: | 2008-02-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Meriolins, a new class of cell death inducing kinase inhibitors with enhanced selectivity for cyclin-dependent kinases
Cancer Res., 67, 2007
|
|
8HJB
 
 | | Crystal structure of Pseudomonas aeruginosa PvrA with coenzyme A | | Descriptor: | COENZYME A, TetR family transcriptional regulator | | Authors: | Liang, H, Bartlam, M. | | Deposit date: | 2022-11-23 | | Release date: | 2023-02-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa.
Nucleic Acids Res., 51, 2023
|
|
6R7F
 
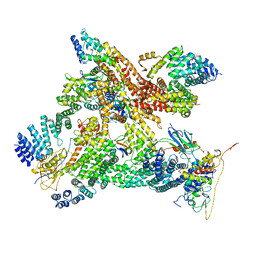 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Martens, C, Ahdash, Z, Yebenes, H, Schmidt, C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
5M23
 
 | | Modulation of MLL1 Methyltransferase Activity | | Descriptor: | 4-[(~{E})-[4-[[[(2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-[[(2~{S})-2-azanyl-3-oxidanyl-propanoyl]amino]propanoyl]amino]-5-carbamimidamido-pentanoyl]amino]propanoyl]amino]methyl]phenyl]diazenyl]-~{N}-[(2~{S})-3-methyl-1-oxidanylidene-butan-2-yl]benzamide, WD repeat-containing protein 5 | | Authors: | Srinivasan, V. | | Deposit date: | 2016-10-11 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Controlled inhibition of methyltransferases using photoswitchable peptidomimetics: towards an epigenetic regulation of leukemia.
Chem Sci, 8, 2017
|
|
6R7H
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.V, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6R7I
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Faull, S.F, Lau, A.M.C, Beuron, F, Cronin, N.B, Morris, E.P, Politis, A. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
