6M2C
 
 | |
4WHV
 
 | | E3 ubiquitin-protein ligase RNF8 in complex with Ubiquitin-conjugating enzyme E2 N and Polyubiquitin-B | | Descriptor: | E3 ubiquitin-protein ligase RNF8, Polyubiquitin-B, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-09-23 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (8.3 Å) | | Cite: | RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J.Biol.Chem., 291, 2016
|
|
4LDT
 
 | | The structure of h/ceOTUB1-ubiquitin aldehyde-UBCH5B~Ub | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Ubiquitin, ... | | Authors: | Wiener, R, DiBello, A.T, Lombardi, P.M, Guzzo, C.M, Zhang, X, Matunis, M.J, Wolberger, C. | | Deposit date: | 2013-06-25 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1.
Nat.Struct.Mol.Biol., 20, 2013
|
|
5NKZ
 
 | | Crystal structure of H. polymorpha ubiquitin conjugating enzyme Pex4p in complex with soluble domain of Pex22p | | Descriptor: | Peroxin 22, Ubiquitin-conjugating enzyme E2-21 kDa | | Authors: | Danda, N, Lunev, S, Ali, A, Groves, M.R, Williams, C. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into K48-linked ubiquitin chain formation by the Pex4p-Pex22p complex.
Biochem. Biophys. Res. Commun., 496, 2018
|
|
4WZ3
 
 | | Crystal structure of the complex between LubX/LegU2/Lpp2887 U-box 1 and Homo sapiens UBE2D2 | | Descriptor: | E3 ubiquitin-protein ligase LubX, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Stogios, P.J, Quaile, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
4LAD
 
 | | Crystal Structure of the Ube2g2:RING-G2BR complex | | Descriptor: | E3 ubiquitin-protein ligase AMFR, OXALATE ION, Ubiquitin-conjugating enzyme E2 G2, ... | | Authors: | Liang, Y.-H, Li, J, Das, R, Byrd, R.A, Ji, X. | | Deposit date: | 2013-06-19 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
7K5J
 
 | | Structure of an E1-E2-ubiquitin thioester mimetic | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ubiquitin, Ubiquitin-activating enzyme E1 1, ... | | Authors: | Yuan, L, Lv, Z, Olsen, S.K. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Crystal structures of an E1-E2-ubiquitin thioester mimetic reveal molecular mechanisms of transthioesterification.
Nat Commun, 12, 2021
|
|
4X57
 
 | |
6N13
 
 | | UbcH7-Ub Complex with R0RBR Parkin and phosphoubiquitin | | Descriptor: | E3 ubiquitin-protein ligase parkin, Ubiquitin-conjugating enzyme E2 L3, ZINC ION, ... | | Authors: | Condos, T.E.C, Dunkerley, K.M, Freeman, E.A, Barber, K.R, Aguirre, J.D, Chaugule, V.K, Xiao, Y, Konermann, L, Walden, H, Shaw, G.S. | | Deposit date: | 2018-11-08 | | Release date: | 2018-11-28 | | Last modified: | 2020-01-08 | | Method: | SOLUTION NMR | | Cite: | Synergistic recruitment of UbcH7~Ub and phosphorylated Ubl domain triggers parkin activation.
EMBO J., 37, 2018
|
|
6NYA
 
 | | Crystal Structure of ubiquitin E1 (Uba1) in complex with Ubc3 (Cdc34) and ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Olsen, S.K, Williams, K.M, Atkison, J.H. | | Deposit date: | 2019-02-11 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.065 Å) | | Cite: | Structural insights into E1 recognition and the ubiquitin-conjugating activity of the E2 enzyme Cdc34.
Nat Commun, 10, 2019
|
|
6NYO
 
 | | Crystal structure of a human Cdc34-ubiquitin thioester mimetic | | Descriptor: | 1,2-ETHANEDIOL, 4,5-dideoxy-5-(3',5'-dichlorobiphenyl-4-yl)-4-[(methoxyacetyl)amino]-L-arabinonic acid, PHOSPHATE ION, ... | | Authors: | Olsen, S.K, Williams, K.M, Atkison, J.H. | | Deposit date: | 2019-02-11 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Structural insights into E1 recognition and the ubiquitin-conjugating activity of the E2 enzyme Cdc34.
Nat Commun, 10, 2019
|
|
4MDK
 
 | | Cdc34-ubiquitin-CC0651 complex | | Descriptor: | 4,5-dideoxy-5-(3',5'-dichlorobiphenyl-4-yl)-4-[(methoxyacetyl)amino]-L-arabinonic acid, Ubiquitin, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, Orlicky, S, Tyers, M, Sicheri, F. | | Deposit date: | 2013-08-22 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6095 Å) | | Cite: | E2 enzyme inhibition by stabilization of a low-affinity interface with ubiquitin.
Nat.Chem.Biol., 10, 2014
|
|
7KZV
 
 | |
7KZT
 
 | |
7KZR
 
 | | Structure of the human Fanconi Anaemia Core-UBE2T-ID complex | | Descriptor: | E3 ubiquitin-protein ligase FANCL, Fanconi anemia core complex-associated protein 100, Fanconi anemia core complex-associated protein 20, ... | | Authors: | Wang, S.L, Pavletich, N.P. | | Deposit date: | 2020-12-10 | | Release date: | 2021-03-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of the FA core ubiquitin ligase closing the ID clamp on DNA.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KZS
 
 | |
3E46
 
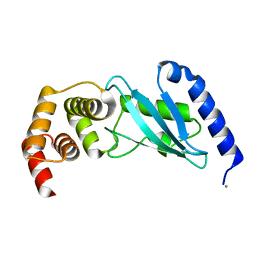 | | Crystal structure of ubiquitin-conjugating enzyme E2-25kDa (Huntington interacting protein 2) M172A mutant | | Descriptor: | CALCIUM ION, Ubiquitin-conjugating enzyme E2-25 kDa | | Authors: | Hughes, R.C, Wilson, R.C, Flatt, J.W, Meehan, E.J, Ng, J.D, Twigg, P.D. | | Deposit date: | 2008-08-09 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of full-length ubiquitin-conjugating enzyme E2-25K (huntingtin-interacting protein 2).
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3EB6
 
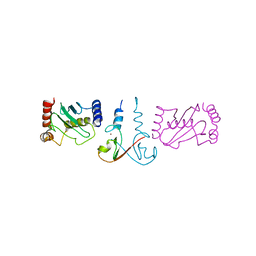 | | Structure of the cIAP2 RING domain bound to UbcH5b | | Descriptor: | Baculoviral IAP repeat-containing protein 3, Ubiquitin-conjugating enzyme E2 D2, ZINC ION | | Authors: | Mace, P.D, Linke, K, Schumacher, F.-R, Smith, C.A, Day, C.L. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of the cIAP2 RING Domain Reveal Conformational Changes Associated with Ubiquitin-conjugating Enzyme (E2) Recruitment.
J.Biol.Chem., 283, 2008
|
|
3HCU
 
 | | Crystal structure of TRAF6 in complex with Ubc13 in the C2 space group | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin-conjugating enzyme E2 N, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3HCT
 
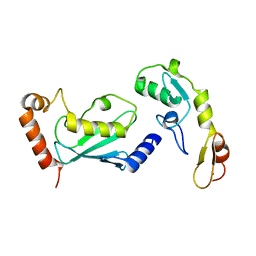 | | Crystal structure of TRAF6 in complex with Ubc13 in the P1 space group | | Descriptor: | TNF receptor-associated factor 6, Ubiquitin-conjugating enzyme E2 N, ZINC ION | | Authors: | Yin, Q, Lin, S.-C, Lamothe, B, Lu, M, Lo, Y.-C, Hura, G, Zheng, L, Rich, R.L, Campos, A.D, Myszka, D.G, Lenardo, M.J, Darnay, B.G, Wu, H. | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | E2 interaction and dimerization in the crystal structure of TRAF6.
Nat.Struct.Mol.Biol., 16, 2009
|
|
7M2K
 
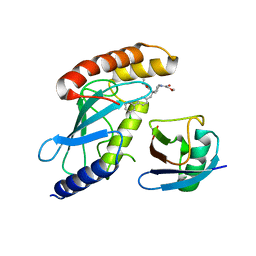 | | CDC34A-Ubiquitin-2ab inhibitor complex | | Descriptor: | 4-[(3',5'-dichloro[1,1'-biphenyl]-4-yl)methyl]-N-ethyl-1-(methoxyacetyl)piperidine-4-carboxamide, Ubiquitin, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, St-Cyr, D, Tyers, M, Sicheri, F. | | Deposit date: | 2021-03-16 | | Release date: | 2021-11-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Identification and optimization of molecular glue compounds that inhibit a noncovalent E2 enzyme-ubiquitin complex.
Sci Adv, 7, 2021
|
|
7MEX
 
 | | Structure of yeast Ubr1 in complex with Ubc2 and N-degron | | Descriptor: | E3 ubiquitin-protein ligase UBR1, N-degron, Ubiquitin, ... | | Authors: | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
7MEY
 
 | | Structure of yeast Ubr1 in complex with Ubc2 and monoubiquitinated N-degron | | Descriptor: | 2-(ethylamino)ethane-1-thiol, E3 ubiquitin-protein ligase UBR1, Monoubiquitinated N-degron, ... | | Authors: | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-11-24 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
3PTF
 
 | |
3FN1
 
 | | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. | | Descriptor: | NEDD8-activating enzyme E1 catalytic subunit, NEDD8-conjugating enzyme UBE2F | | Authors: | Huang, D.T, Ayrault, O, Hunt, H.W, Taherbhoy, A.M, Duda, D.M, Scott, D.C, Borg, L.A, Neale, G, Murray, P.J, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2008-12-22 | | Release date: | 2009-03-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification
Mol.Cell, 33, 2009
|
|
