3BF6
 
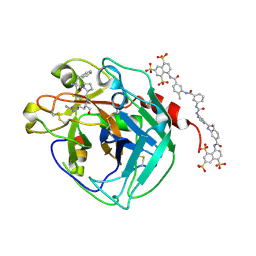 | | Thrombin:suramin complex | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFONIC ACID, PHE-PRO-ARG, Thrombin, ... | | Authors: | Lima, L.M.T.R, Polikarpov, I, Monteiro, R.Q. | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-25 | | Last modified: | 2015-07-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and thermodynamic analysis of thrombin:suramin interaction in solution and crystal phases.
Biochim.Biophys.Acta, 1794, 2009
|
|
1CXL
 
 | |
1TGB
 
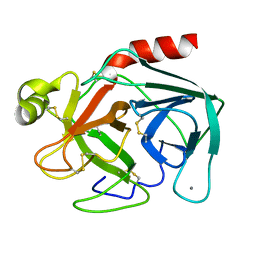 | | CRYSTAL STRUCTURE OF BOVINE TRYPSINOGEN AT 1.8 ANGSTROMS RESOLUTION. II. CRYSTALLOGRAPHIC REFINEMENT, REFINED CRYSTAL STRUCTURE AND COMPARISON WITH BOVINE TRYPSIN | | Descriptor: | CALCIUM ION, TRYPSINOGEN | | Authors: | Bode, W, Fehlhammer, H, Huber, R. | | Deposit date: | 1979-03-07 | | Release date: | 1979-06-13 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of bovine trypsinogen at 1-8 A resolution. II. Crystallographic refinement, refined crystal structure and comparison with bovine trypsin.
J.Mol.Biol., 111, 1977
|
|
2AWW
 
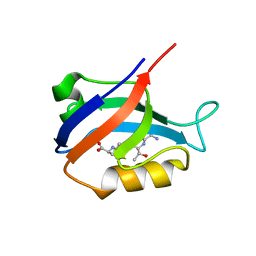 | | Synapse associated protein 97 PDZ2 domain variant C378G with C-terminal GluR-A peptide | | Descriptor: | 18-residue C-terminal peptide from glutamate receptor, ionotropic, AMPA1, ... | | Authors: | Von Ossowski, I, Oksanen, E, Von Ossowski, L, Cai, C, Sundberg, M, Goldman, A, Keinanen, K. | | Deposit date: | 2005-09-02 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the second PDZ domain of SAP97 in complex with a GluR-A C-terminal peptide
Febs J., 273, 2006
|
|
4Q94
 
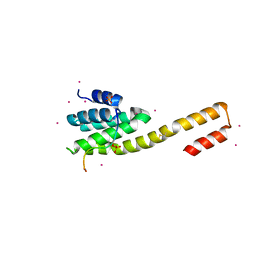 | | human RPRD1B CID in complex with a RPB1-CTD derived Ser2 phosphorylated peptide | | Descriptor: | Regulation of nuclear pre-mRNA domain-containing protein 1B, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-04-29 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3NSZ
 
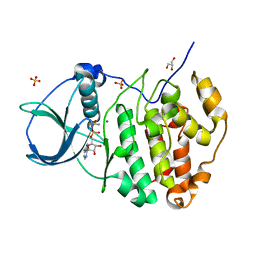 | | Human CK2 catalytic domain in complex with AMPPN | | Descriptor: | Casein kinase II subunit alpha, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of CX-4945 binding to human protein kinase CK2.
Febs Lett., 585, 2011
|
|
4PU9
 
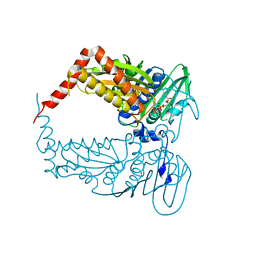 | |
1ERI
 
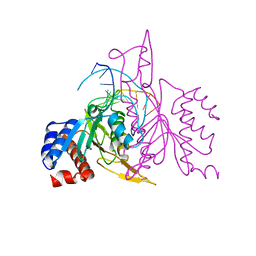 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE-DNA RECOGNITION COMPLEX: THE RECOGNITION NETWORK AND THE INTEGRATION OF RECOGNITION AND CLEAVAGE | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PROTEIN (ECO RI ENDONUCLEASE (E.C.3.1.21.4)) | | Authors: | Kim, Y, Grable, J.C, Love, R, Greene, P.J, Rosenberg, J.M. | | Deposit date: | 1994-05-18 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing.
Science, 249, 1990
|
|
4PJ3
 
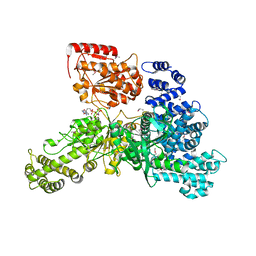 | | Structural insight into the function and evolution of the spliceosomal helicase Aquarius, Structure of Aquarius in complex with AMPPNP | | Descriptor: | Intron-binding protein aquarius, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | De, I, Bessonov, S, Hofele, R, dos Santos, K.F, Will, C.L, Urlaub, H, Luhrmann, R, Pena, V. | | Deposit date: | 2014-05-11 | | Release date: | 2015-01-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The RNA helicase Aquarius exhibits structural adaptations mediating its recruitment to spliceosomes.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1ES2
 
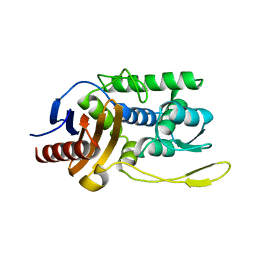 | | S96A mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
2ZVM
 
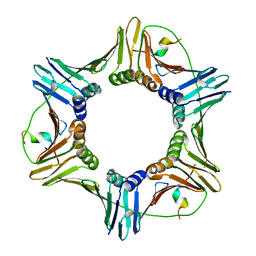 | | Crystal structure of PCNA in complex with DNA polymerase iota fragment | | Descriptor: | DNA polymerase iota, Proliferating cell nuclear antigen | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
1E5Z
 
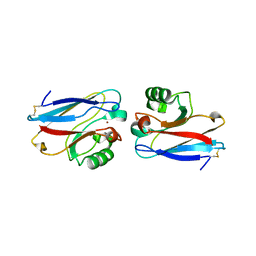 | |
4PGM
 
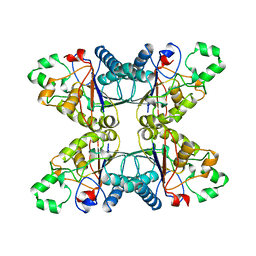 | |
3JAJ
 
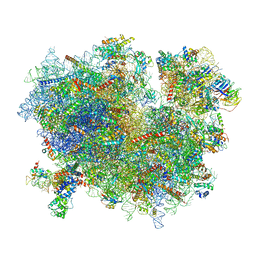 | |
1TGT
 
 | | ON THE DISORDERED ACTIVATION DOMAIN IN TRYPSINOGEN. CHEMICAL LABELLING AND LOW-TEMPERATURE CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, METHANOL, TRYPSINOGEN | | Authors: | Walter, J, Steigemann, W, Singh, T.P, Bartunik, H, Bode, W, Huber, R. | | Deposit date: | 1981-10-26 | | Release date: | 1982-03-04 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | On the Disordered Activation Domain in Trypsinogen. Chemical Labelling and Low-Temperature Crystallography
Acta Crystallogr.,Sect.B, 38, 1982
|
|
1ES3
 
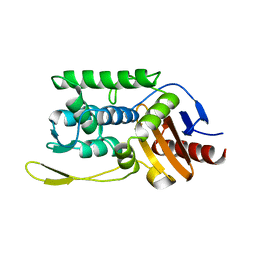 | | C98A mutant of streptomyces K15 DD-transpeptidase | | Descriptor: | DD-TRANSPEPTIDASE, SODIUM ION | | Authors: | Fonze, E, Charlier, P. | | Deposit date: | 2000-04-07 | | Release date: | 2000-05-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic mechanism of the Streptomyces K15 DD-transpeptidase/penicillin-binding protein probed by site-directed mutagenesis and structural analysis.
Biochemistry, 42, 2003
|
|
2ZR1
 
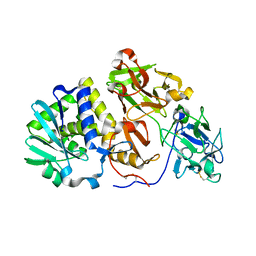 | | Agglutinin from Abrus Precatorius | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Agglutinin-1 chain A, Agglutinin-1 chain B | | Authors: | Cheng, J, Lu, T.H, Liu, C.L, Lin, J.Y. | | Deposit date: | 2008-08-22 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A biophysical elucidation for less toxicity of Agglutinin than Abrin-a from the Seeds of Abrus Precatorius in consequence of crystal structure
J.Biomed.Sci., 17, 2010
|
|
2B97
 
 | | Ultra-high resolution structure of hydrophobin HFBII | | Descriptor: | Hydrophobin II, MANGANESE (II) ION | | Authors: | Hakanpaa, J, Linder, M, Popov, A, Schmidt, A, Rouvinen, J. | | Deposit date: | 2005-10-11 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Hydrophobin HFBII in detail: ultrahigh-resolution structure at 0.75 A.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4PS0
 
 | |
4PRV
 
 | |
4PU5
 
 | | Shewanella oneidensis Toxin Antitoxin System Toxin Protein HipA Bound with AMPPNP and Mg | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Wen, Y, Behiels, E, Felix, J, Elegheert, J, Vergauwen, B, Devreese, B, Savvides, S. | | Deposit date: | 2014-03-12 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | The bacterial antitoxin HipB establishes a ternary complex with operator DNA and phosphorylated toxin HipA to regulate bacterial persistence.
Nucleic Acids Res., 42, 2014
|
|
2BCU
 
 | | DNA polymerase lambda in complex with a DNA duplex containing an unpaired Damp and a T:T mismatch | | Descriptor: | 5'-D(*CP*AP*GP*TP*TP*CP*G)-3', 5'-D(*CP*GP*GP*CP*CP*GP*AP*TP*AP*CP*TP*G)-3', 5'-D(P*GP*CP*CP*G)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2005-10-19 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase
Cell(Cambridge,Mass.), 124, 2006
|
|
1V8K
 
 | | The Crystal Structure of the Minimal Functional Domain of the Microtubule Destabilizer KIF2C Complexed with Mg-AMPPNP | | Descriptor: | Kinesin-like protein KIF2C, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Ogawa, T, Nitta, R, Okada, Y, Hirokawa, N. | | Deposit date: | 2004-01-09 | | Release date: | 2004-03-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A common mechanism for microtubule destabilizers-M type kinesins stabilize curling of the protofilament using the class-specific neck and loops.
Cell(Cambridge,Mass.), 116, 2004
|
|
2AMD
 
 | | Crystal Structure Of SARS_CoV Mpro in Complex with an Inhibitor N9 | | Descriptor: | 3C-like proteinase, N-(3-FUROYL)-D-VALYL-L-VALYL-N~1~-((1R,2Z)-4-ETHOXY-4-OXO-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-D-LEUCINAMIDE | | Authors: | Yang, H, Xue, X, Yang, K, Zhao, Q, Bartlam, M, Rao, Z. | | Deposit date: | 2005-08-09 | | Release date: | 2005-09-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design of Wide-Spectrum Inhibitors Targeting Coronavirus Main Proteases.
Plos Biol., 3, 2005
|
|
4Q3K
 
 | | Crystal structure of MGS-M1, an alpha/beta hydrolase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | CHLORIDE ION, FLUORIDE ION, MGS-M1, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
