1TLD
 
 | | CRYSTAL STRUCTURE OF BOVINE BETA-TRYPSIN AT 1.5 ANGSTROMS RESOLUTION IN A CRYSTAL FORM WITH LOW MOLECULAR PACKING DENSITY. ACTIVE SITE GEOMETRY, ION PAIRS AND SOLVENT STRUCTURE | | Descriptor: | BETA-TRYPSIN, CALCIUM ION, SULFATE ION | | Authors: | Bartunik, H.D, Summers, L.J, Bartsch, H.H. | | Deposit date: | 1989-07-24 | | Release date: | 1990-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of bovine beta-trypsin at 1.5 A resolution in a crystal form with low molecular packing density. Active site geometry, ion pairs and solvent structure.
J.Mol.Biol., 210, 1989
|
|
2AU7
 
 | | The R43Q active site variant of E.coli inorganic pyrophosphatase | | Descriptor: | CHLORIDE ION, Inorganic pyrophosphatase, MANGANESE (II) ION, ... | | Authors: | Samygina, V.R, Avaeva, S.M, Bartunik, H.D. | | Deposit date: | 2005-08-27 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Reversible inhibition of Escherichia coli inorganic pyrophosphatase by fluoride: trapped catalytic intermediates in cryo-crystallographic studies
J.Mol.Biol., 366, 2007
|
|
2AUU
 
 | | Inorganic pyrophosphatase complexed with magnesium pyrophosphate and fluoride | | Descriptor: | CHLORIDE ION, FLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Samygina, V.R, Popov, A.N, Avaeva, S.M, Bartunik, H.D. | | Deposit date: | 2005-08-29 | | Release date: | 2006-08-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Reversible inhibition of Escherichia coli inorganic pyrophosphatase by fluoride: trapped catalytic intermediates in cryo-crystallographic studies
J.Mol.Biol., 366, 2007
|
|
1QV9
 
 | | Coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) from Methanopyrus kandleri: A methanogenic enzyme with an unusual quarternary structure | | Descriptor: | F420-dependent methylenetetrahydromethanopterin dehydrogenase, MAGNESIUM ION | | Authors: | Hagemeier, C.H, Shima, S, Thauer, R.K, Bourenkov, G, Bartunik, H.D, Ermler, U. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Coenzyme F420-dependent methylenetetrahydromethanopterin dehydrogenase (Mtd) from Methanopyrus kandleri: a methanogenic enzyme with an unusual quarternary structure
J.Mol.Biol., 332, 2003
|
|
3K7T
 
 | | Crystal structure of apo-form 6-hydroxy-L-nicotine oxidase, crystal form P3121 | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Bourenkov, G.P, Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2009-10-13 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure Analysis of Free and Substrate-Bound 6-Hydroxy-l-Nicotine Oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3K7M
 
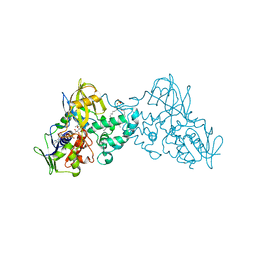 | | Crystal structure of 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Bourenkov, G.P, Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2009-10-13 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure Analysis of Free and Substrate-Bound 6-Hydroxy-l-Nicotine Oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3K7Q
 
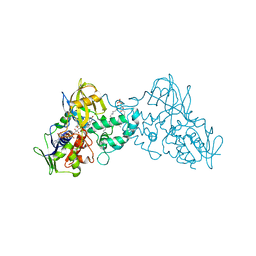 | | Crystal structure of substrate-bound 6-hydroxy-L-nicotine oxidase from Arthrobacter nicotinovorans | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 5-[(2S)-1-methylpyrrolidin-2-yl]pyridin-2-ol, 6-hydroxy-L-nicotine oxidase, ... | | Authors: | Bourenkov, G.P, Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2009-10-13 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure Analysis of Free and Substrate-Bound 6-Hydroxy-l-Nicotine Oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
3SS7
 
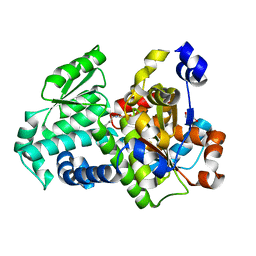 | | Crystal structure of holo D-serine dehydratase from Escherichia coli at 1.55 A resolution | | Descriptor: | D-serine dehydratase, GLYCEROL, POTASSIUM ION, ... | | Authors: | Urusova, D.V, Isupov, M.N, Antonyuk, S.V, Kachalova, G.S, Vagin, A.A, Lebedev, A.A, Bourenkov, G.P, Dauter, Z, Bartunik, H.D, Melik-Adamyan, W.R, Mueller, T.D, Schnackerz, K.D. | | Deposit date: | 2011-07-07 | | Release date: | 2012-01-18 | | Last modified: | 2012-02-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure of D-serine dehydratase from Escherichia coli.
Biochim.Biophys.Acta, 1824, 2011
|
|
3SS9
 
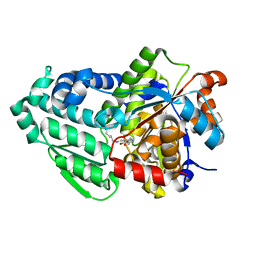 | | Crystal structure of holo D-serine dehydratase from Escherichia coli at 1.97 A resolution | | Descriptor: | D-serine dehydratase, POTASSIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Urusova, D.V, Isupov, M.N, Antonyuk, S.V, Kachalova, G.S, Vagin, A.A, Lebedev, A.A, Bourenkov, G.P, Dauter, Z, Bartunik, H.D, Melik-Adamyan, W.R, Mueller, T.D, Schnackerz, K.D. | | Deposit date: | 2011-07-08 | | Release date: | 2012-01-18 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of D-serine dehydratase from Escherichia coli.
Biochim.Biophys.Acta, 1824, 2011
|
|
5LHC
 
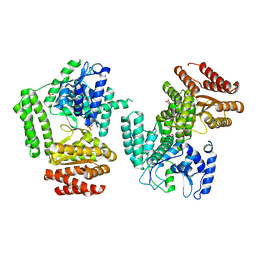 | | The structure of D456A mutant of Nt.BspD6I nicking endonuclease at 0.24 nm resolution . | | Descriptor: | GLYCEROL, Nicking endonuclease N.BspD6I, PHOSPHATE ION | | Authors: | Kachalova, G.S, Yunusova, A.K, Popov, A.N, Artyukh, R.I, Perevyazova, T.A, Bartunik, H.D, Zheleznaya, L.A. | | Deposit date: | 2016-07-10 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural implication of activity loss by D456A mutant of the nicking endonuclease Nt.BspD6I.
To Be Published
|
|
5LIQ
 
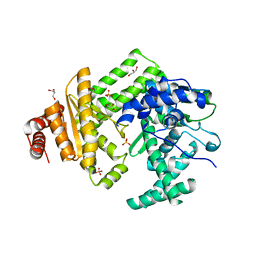 | | The structure of C160S,C508S,C578S mutant of Nt.BspD6I nicking endonuclease at 0.185 nm resolution . | | Descriptor: | GLYCEROL, Nicking endonuclease N.BspD6I, PHOSPHATE ION | | Authors: | Kachalova, G.S, Artyukh, R.I, Perevyazova, T.A, Yunusova, A.K, Popov, A.N, Bartunik, H.D, Zheleznaya, L.A. | | Deposit date: | 2016-07-15 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural features of Cysteine residues mutation of the nicking endonuclease Nt.BspD6I.
To Be Published
|
|
1C52
 
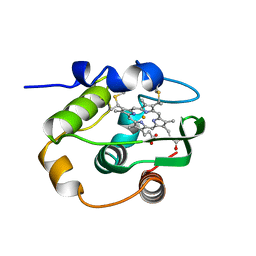 | | THERMUS THERMOPHILUS CYTOCHROME-C552: A NEW HIGHLY THERMOSTABLE CYTOCHROME-C STRUCTURE OBTAINED BY MAD PHASING | | Descriptor: | CYTOCHROME-C552, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Than, M.E, Hof, P, Huber, R, Bourenkov, G.P, Bartunik, H.D, Buse, G, Soulimane, T. | | Deposit date: | 1997-06-23 | | Release date: | 1998-06-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Thermus thermophilus cytochrome-c552: A new highly thermostable cytochrome-c structure obtained by MAD phasing.
J.Mol.Biol., 271, 1997
|
|
5LIZ
 
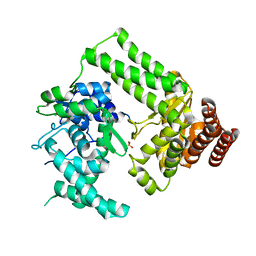 | | The structure of Nt.BspD6I nicking endonuclease with all cysteines mutated by serine residues at 0.19 nm resolution . | | Descriptor: | Nicking endonuclease N.BspD6I, PHOSPHATE ION | | Authors: | Kachalova, G.S, Artyukh, R.I, Perevyazova, T.A, Yunusova, A.K, Popov, A.N, Bartunik, H.D, Zheleznaya, L.A. | | Deposit date: | 2016-07-16 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural features of Cysteine residues mutation of the nicking endonuclease Nt.BspD6I.
To Be Published
|
|
2QHF
 
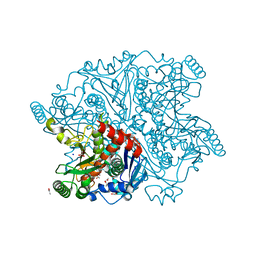 | | Mycobacterium tuberculosis Chorismate synthase in complex with NCA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Bruning, M, Bourenkov, G.P, Strizhov, N.I, Bartunik, H.D. | | Deposit date: | 2007-07-02 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mycobacterium tuberculosis Chorismate synthase in complex with NCA
To be Published
|
|
2TGA
 
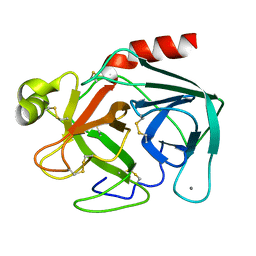 | | ON THE DISORDERED ACTIVATION DOMAIN IN TRYPSINOGEN. CHEMICAL LABELLING AND LOW-TEMPERATURE CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, TRYPSINOGEN | | Authors: | Walter, J, Steigemann, W, Singh, T.P, Bartunik, H, Bode, W, Huber, R. | | Deposit date: | 1981-10-26 | | Release date: | 1982-03-04 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | On the Disordered Activation Domain in Trypsinogen. Chemical Labelling and Low-Temperature Crystallography
Acta Crystallogr.,Sect.B, 38, 1982
|
|
2TGT
 
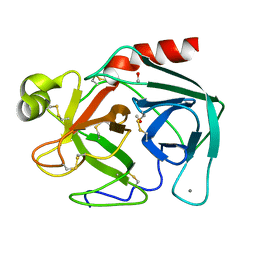 | | ON THE DISORDERED ACTIVATION DOMAIN IN TRYPSINOGEN. CHEMICAL LABELLING AND LOW-TEMPERATURE CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, METHANOL, TRYPSINOGEN | | Authors: | Walter, J, Steigemann, W, Singh, T.P, Bartunik, H, Bode, W, Huber, R. | | Deposit date: | 1981-10-26 | | Release date: | 1982-03-04 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | On the Disordered Activation Domain in Trypsinogen. Chemical Labelling and Low-Temperature Crystallography
Acta Crystallogr.,Sect.B, 38, 1982
|
|
3CFF
 
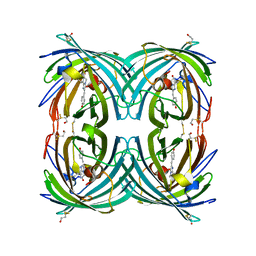 | |
3CFH
 
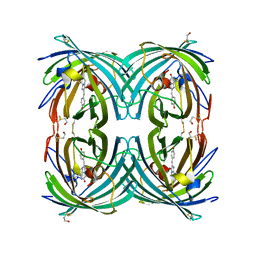 | |
3CFA
 
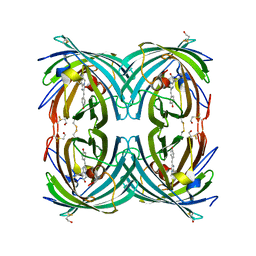 | |
2EWF
 
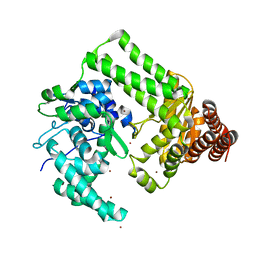 | | Crystal structure of the site-specific DNA nickase N.BspD6I | | Descriptor: | BROMIDE ION, Nicking endonuclease N.BspD6I | | Authors: | Kachalova, G.S, Bartunik, H.D, Artyukh, R.I, Rogulin, E.A, Perevyazova, T.A, Zheleznaya, L.A, Matvienko, N.I. | | Deposit date: | 2005-11-03 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural analysis of the heterodimeric type IIS restriction endonuclease R.BspD6I acting as a complex between a monomeric site-specific nickase and a catalytic subunit.
J.Mol.Biol., 384, 2008
|
|
3FZN
 
 | | Intermediate analogue in benzoylformate decarboxylase | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(S)-hydroxy[(R)-hydroxy(methoxy)phosphoryl]phenylmethyl}-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, CHLORIDE ION, ... | | Authors: | Bruning, M, Berheide, M, Meyer, D, Golbik, R, Bartunik, H, Liese, A, Tittmann, K. | | Deposit date: | 2009-01-26 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural and kinetic studies on native intermediates and an intermediate analogue in benzoylformate decarboxylase reveal a least motion mechanism with an unprecedented short-lived predecarboxylation intermediate.
Biochemistry, 48, 2009
|
|
1VET
 
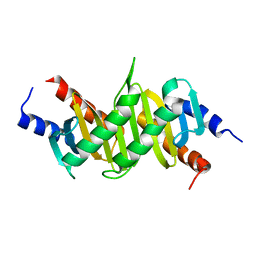 | | Crystal Structure of p14/MP1 at 1.9 A resolution | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein, Mitogen-activated protein kinase kinase 1 interacting protein 1 | | Authors: | Kurzbauer, R, Teis, D, Maurer-Stroh, S, Eisenhaber, F, Hekman, M, Bourenkov, G.P, Bartunik, H.D, Huber, L.A, Clausen, T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the p14/MP1 scaffolding complex: How a twin couple attaches mitogen- activated protein kinase signaling to late endosomes
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1VEU
 
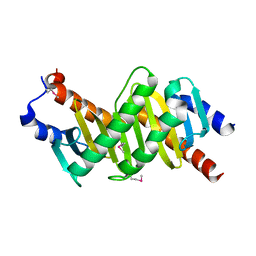 | | Crystal structure of the p14/MP1 complex at 2.15 A resolution | | Descriptor: | Late endosomal/lysosomal Mp1 interacting protein, Mitogen-activated protein kinase kinase 1 interacting protein 1 | | Authors: | Kurzbauer, R, Teis, D, Maurer-Stroh, S, Eisenhaber, F, Hekman, M, Bourenkov, G.P, Bartunik, H.D, Huber, L.A, Clausen, T. | | Deposit date: | 2004-04-05 | | Release date: | 2004-08-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the p14/MP1 scaffolding complex: How a twin couple attaches mitogen- activated protein kinase signaling to late endosomes
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1KFX
 
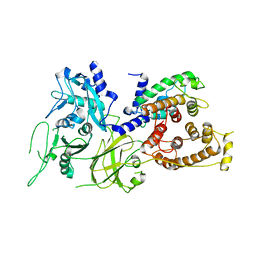 | | Crystal Structure of Human m-Calpain Form I | | Descriptor: | M-CALPAIN LARGE SUBUNIT, M-CALPAIN SMALL SUBUNIT | | Authors: | Strobl, S, Fernandez-Catalan, C, Braun, M, Huber, R, Masumoto, H, Nakagawa, K, Irie, A, Sorimachi, H, Bourenkow, G, Bartunik, H, Suzuki, K, Bode, W. | | Deposit date: | 2001-11-23 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1KFU
 
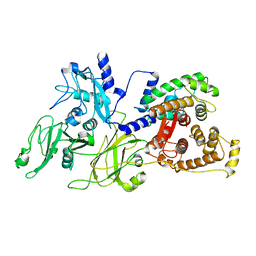 | | Crystal Structure of Human m-Calpain Form II | | Descriptor: | M-CALPAIN LARGE SUBUNIT, M-CALPAIN SMALL SUBUNIT | | Authors: | Strobl, S, Fernandez-Catalan, C, Braun, M, Huber, R, Masumoto, H, Nakagawa, K, Irie, A, Sorimachi, H, Bourenkow, G, Bartunik, H, Suzuki, K, Bode, W. | | Deposit date: | 2001-11-23 | | Release date: | 2001-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of calcium-free human m-calpain suggests an electrostatic switch mechanism for activation by calcium.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
