4PK6
 
 | | Crystal structure of the indoleamine 2,3-dioxygenagse 1 (IDO1) complexed with imidazothiazole derivative | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-[2-(3-chlorophenyl)ethyl]-3-(4-methylphenyl)imidazo[2,1-b][1,3]thiazole-5-carboxamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kohno, T, Tojo, S, Ishii, T, Kamioka, S. | | Deposit date: | 2014-05-13 | | Release date: | 2014-09-03 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Crystal Structures and Structure-Activity Relationships of Imidazothiazole Derivatives as IDO1 Inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
4PP9
 
 | |
6E5T
 
 | | Crystal structure of human cellular retinol binding protein 1 in complex with abnormal-cannabidiorcin (Abn-CBDO) | | Descriptor: | (1'R,2'R)-5',6-dimethyl-2'-(prop-1-en-2-yl)-1',2',3',4'-tetrahydro[1,1'-biphenyl]-2,4-diol, Retinol-binding protein 1 | | Authors: | Silvaroli, J.A, Horwitz, S, Banerjee, S, Kiser, P.D, Golczak, M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Abnormal Cannabidiol Modulates Vitamin A Metabolism by Acting as a Competitive Inhibitor of CRBP1.
Acs Chem.Biol., 14, 2019
|
|
4P6W
 
 | | Crystal Structure of mometasone furoate-bound glucocorticoid receptor ligand binding domain | | Descriptor: | Glucocorticoid receptor, MOMETASONE FUROATE, Nuclear receptor coactivator 2 | | Authors: | He, Y, Zhou, X.E, Tolbert, W.D, Powell, K, Melcher, K, Xu, H.E. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structures and mechanism for the design of highly potent glucocorticoids.
Cell Res., 24, 2014
|
|
4PA0
 
 | | Omecamtiv Mercarbil binding site on the Human Beta-Cardiac Myosin Motor Domain | | Descriptor: | GLYCEROL, Myosin-7,Green fluorescent protein, methyl 4-(2-fluoro-3-{[(6-methylpyridin-3-yl)carbamoyl]amino}benzyl)piperazine-1-carboxylate | | Authors: | Winkelmann, D.A, Miller, M.T, Stock, A.M. | | Deposit date: | 2014-04-06 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for drug-induced allosteric changes to human beta-cardiac myosin motor activity.
Nat Commun, 6, 2015
|
|
7CGW
 
 | | Complex structure of PD-1 and tislelizumab Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of tislelizumab Fab, Light chain of tislelizumab Fab, ... | | Authors: | Hong, Y, Feng, Y.C, Liu, Y. | | Deposit date: | 2020-07-02 | | Release date: | 2021-04-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Tislelizumab uniquely binds to the CC' loop of PD-1 with slow-dissociated rate and complete PD-L1 blockage.
Febs Open Bio, 11, 2021
|
|
4CLJ
 
 | | Structure of L1196M Mutant Human Anaplastic Lymphoma Kinase in Complex with PF-06463922 ((10R)-7-amino-12-fluoro-2,10,16-trimethyl- 15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11) benzoxadiazacyclotetradecine-3-carbonitrile). | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M.A, Deng, Y.L, Liu, W, Brooun, A, Stewart, A.E. | | Deposit date: | 2014-01-14 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of (10R)-7-Amino-12-Fluoro-2,10,16-Trimethyl-15-Oxo-10,15,16,17-Tetrahydro-2H-8,4-(Metheno)Pyrazolo[4,3-H][2,5,11]Benzoxadiazacyclotetradecine-3-Carbonitrile (Pf-06463922), a Macrocyclic Inhibitor of Alk/Ros1 with Pre-Clinical Brain Exposure and Broad Spectrum Potency Against Alk-Resistant Mutations.
J.Med.Chem., 57, 2014
|
|
4CMG
 
 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | 6-(4-fluorophenyl)-5-(4-methoxyphenyl)-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
7CIN
 
 | |
4O97
 
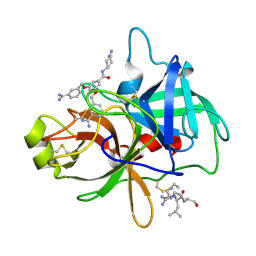 | | Crystal structure of matriptase in complex with inhibitor | | Descriptor: | N-(trans-4-aminocyclohexyl)-3,5-bis[(3-carbamimidoylbenzyl)oxy]benzamide, Peptide CGLR, Suppressor of tumorigenicity 14 protein | | Authors: | Rao, K.N, Chandra, B.R, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S, Subramanya, H.S. | | Deposit date: | 2014-01-02 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided discovery of 1,3,5 tri-substituted benzenes as potent and selective matriptase inhibitors exhibiting in vivo antitumor efficacy.
Bioorg.Med.Chem., 22, 2014
|
|
4CI4
 
 | |
4OIV
 
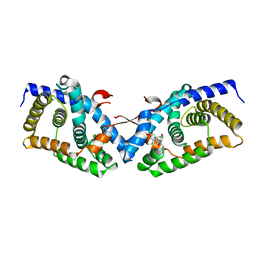 | | Structural basis for small molecule NDB as a selective antagonist of FXR | | Descriptor: | Bile acid receptor, N-benzyl-N-(3-tert-butyl-4-hydroxyphenyl)-2,6-dichloro-4-(dimethylamino)benzamide | | Authors: | Xu, X, Chen, L, Hu, L, Shen, X. | | Deposit date: | 2014-01-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Small Molecule NDB (N-Benzyl-N-(3-(tert-butyl)-4-hydroxyphenyl)-2,6-dichloro-4-(dimethylamino) Benzamide) as a Selective Antagonist of Farnesoid X Receptor alpha (FXR alpha ) in Stabilizing the Homodimerization of the Receptor.
J.Biol.Chem., 290, 2015
|
|
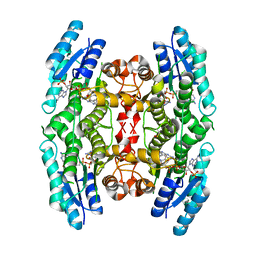
4CLR
 
 | |
4CMC
 
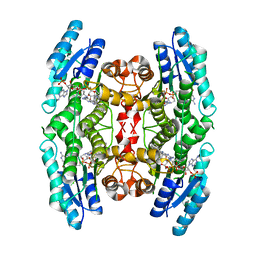 | | Crystal structure of pteridine reductase 1 (PTR1) from Trypanosoma brucei in ternary complex with cofactor and inhibitor | | Descriptor: | N4-cyclohexyl-5,6-diphenyl-7H-pyrrolo[2,3-d]pyrimidine-2,4-diamine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 1 | | Authors: | Barrack, K.L, Hunter, W.N. | | Deposit date: | 2014-01-16 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design and Synthesis of Antiparasitic Pyrrolopyrimidines Targeting Pteridine Reductase 1.
J.Med.Chem., 57, 2014
|
|
4ODT
 
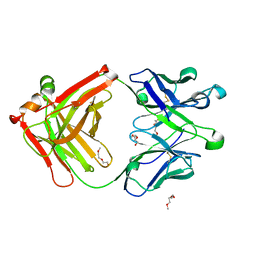 | |
4OH0
 
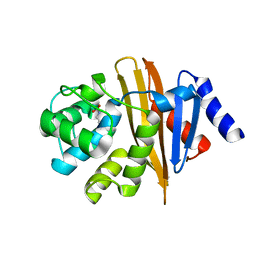 | | Crystal structure of OXA-58 carbapenemase | | Descriptor: | Beta-lactamase OXA-58, CHLORIDE ION | | Authors: | Smith, C.A, Antunes, N.T, Toth, M, Vakulenko, S.B. | | Deposit date: | 2014-01-16 | | Release date: | 2014-02-26 | | Last modified: | 2014-04-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Carbapenemase OXA-58 from Acinetobacter baumannii.
Antimicrob.Agents Chemother., 58, 2014
|
|
4DQL
 
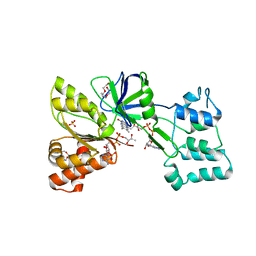 | |
4E1P
 
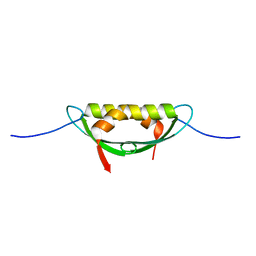 | |
4DLD
 
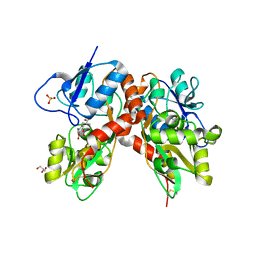 | | Crystal structure of the GluK1 ligand-binding domain (S1S2) in complex with the antagonist (S)-2-amino-3-(2-(2-carboxyethyl)-5-chloro-4-nitrophenyl)propionic acid at 2.0 A resolution | | Descriptor: | (S)-2-amino-3-(2-(2-carboxyethyl)-5-chloro-4-nitrophenyl)propionic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2012-02-06 | | Release date: | 2012-10-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and pharmacological characterization of phenylalanine-based AMPA receptor antagonists at kainate receptors
Chemmedchem, 7, 2012
|
|
4O2A
 
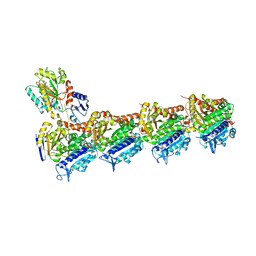 | | Tubulin-BAL27862 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(4-{1-[2-(4-aminophenyl)-2-oxoethyl]-1H-benzimidazol-2-yl}-1,2,5-oxadiazol-3-yl)amino]propanenitrile, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Franck, D, Bachmann, F, Bargsten, K, Buey, R.M, Pohlmann, J, Reinelt, S, Lane, H, Steinmetz, M.O. | | Deposit date: | 2013-12-17 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Novel Microtubule-Destabilizing Drug BAL27862 Binds to the Colchicine Site of Tubulin with Distinct Effects on Microtubule Organization.
J.Mol.Biol., 426, 2014
|
|
7EPU
 
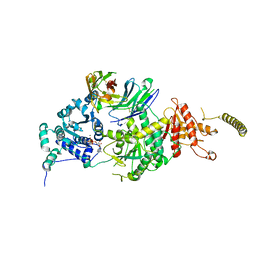 | | Crystal structure of HsALC1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromodomain-helicase-DNA-binding protein 1-like, MAGNESIUM ION, ... | | Authors: | Wang, L, Chen, K.J. | | Deposit date: | 2021-04-27 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of ALC1/CHD1L autoinhibition and the mechanism of activation by the nucleosome.
Nat Commun, 12, 2021
|
|
4DL2
 
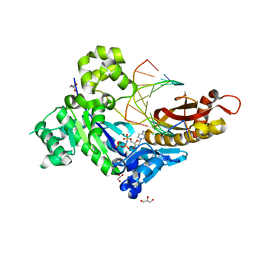 | | Human DNA polymerase eta inserting dCMPNPP opposite CG template (GG0a) | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*CP*GP*GP*CP*TP*CP*AP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*TP*GP*AP*G)-3'), ... | | Authors: | Zhao, Y, Biertumpfel, C, Gregory, M, Hua, Y, Hanaoka, F, Yang, W. | | Deposit date: | 2012-02-05 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for Chemoresistance to Cisplatin Mediated by DNA Polymerase eta
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DOR
 
 | |
4B55
 
 | | Crystal Structure of the Covalent Adduct Formed between Mycobacterium marinum Aryalamine N-acetyltransferase and Phenyl vinyl ketone a derivative of Piperidinols | | Descriptor: | 3-hydroxy-1-phenylpropan-1-one, ARYLAMINE N-ACETYLTRANSFERASE NAT | | Authors: | Abuhammad, A, Fullam, E, Lowe, E.D, Staunton, D, Kawamura, A, Westwood, I.M, Bhakta, S, Garner, A.C, Wilson, D.L, Seden, P.T, Davies, S.G, Russell, A.J, Garman, E.F, Sim, E. | | Deposit date: | 2012-08-02 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Piperidinols that Show Anti-Tubercular Activity as Inhibitors of Arylamine N-Acetyltransferase: An Essential Enzyme for Mycobacterial Survival Inside Macrophages.
Plos One, 7, 2012
|
|
4QMM
 
 | | MST3 IN COMPLEX WITH AT-9283, 4-[(2-{4-[(CYCLOPROPYLCARBAMOYL)AMINO]-1H-PYRAZOL-3-YL}-1H-BENZIMIDAZOL-6-YL)METHYL]MORPHOLIN-4-IUM | | Descriptor: | 1,2-ETHANEDIOL, 1-cyclopropyl-3-{3-[5-(morpholin-4-ylmethyl)-1H-benzimidazol-2-yl]-1H-pyrazol-4-yl}urea, ACETATE ION, ... | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
