7MN6
 
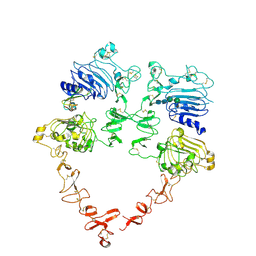 | | Structure of the HER2 S310F/HER3/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Diwanji, D, Trenker, R, Verba, K.A, Jura, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-10-27 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Nature, 600, 2021
|
|
7MN5
 
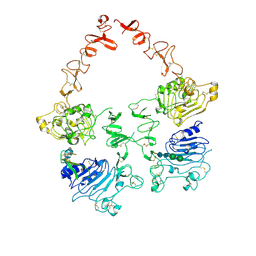 | | Structure of the HER2/HER3/NRG1b Heterodimer Extracellular Domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Diwanji, D, Trenker, R, Verba, K.A, Jura, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-10-27 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Nature, 600, 2021
|
|
7MN8
 
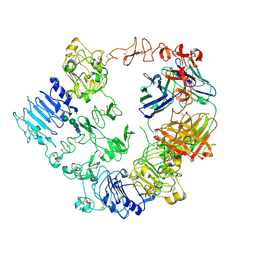 | | Structure of the HER2/HER3/NRG1b Heterodimer Extracellular Domain bound to Trastuzumab Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 6 of Pro-neuregulin-1, ... | | Authors: | Diwanji, D, Trenker, R, Verba, K.A, Jura, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structures of the HER2-HER3-NRG1 beta complex reveal a dynamic dimer interface.
Nature, 600, 2021
|
|
7MK7
 
 | | Augmentor domain of augmentor-beta | | Descriptor: | ALK and LTK ligand 1,Maltodextrin-binding protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krimmer, S.G, Reshetnyak, A.V, Puleo, D.E, Schlessinger, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42815185 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
7M74
 
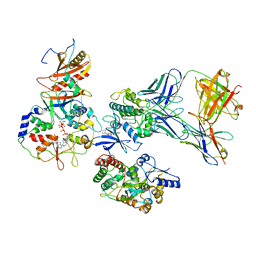 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Mukherjee, S, Harikumar, K.G, Strutzenberg, T, Zhou, X.E, Powell, S.K, Xu, T, Sheldon, R, Lamp, J, Brunzelle, J.S, Radziwon, K, Ellis, A, Novick, S.J, Vega, I.E, Jones, R, Miller, L.J, Xu, H.E, Griffin, P.R, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2021-03-26 | | Release date: | 2021-12-15 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
6DKS
 
 | | Structure of the Rbpj-SHARP-DNA Repressor Complex | | Descriptor: | DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*AP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*TP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Kovall, R.A, VanderWielen, B.D, Yuan, Z. | | Deposit date: | 2018-05-30 | | Release date: | 2019-01-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural and Functional Studies of the RBPJ-SHARP Complex Reveal a Conserved Corepressor Binding Site.
Cell Rep, 26, 2019
|
|
6DBJ
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward stand of RSS signal end, Forward strand of coding flank, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7MHW
 
 | | Crystal structure of the protease inhibitor U-Omp19 from Brucella abortus fused to Maltose-binding protein | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Outer membrane lipoprotein omp19, SULFATE ION | | Authors: | Darriba, M.L, Klinke, S, Otero, L.H, Cerutti, M.L, Cassataro, J, Pasquevich, K.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A disordered region retains the full protease inhibitor activity and the capacity to induce CD8 + T cells in vivo of the oral vaccine adjuvant U-Omp19.
Comput Struct Biotechnol J, 20, 2022
|
|
6D67
 
 | | Crystal structure of the human dual specificity phosphatase 1 catalytic domain (C258S) as a maltose binding protein fusion (maltose bound form) in complex with the designed AR protein mbp3_16 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Designed AR protein mbp3_16, ... | | Authors: | Gumpena, R, Lountos, G.T, Waugh, D.S. | | Deposit date: | 2018-04-20 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | MBP-binding DARPins facilitate the crystallization of an MBP fusion protein.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2RR0
 
 | | Structure of epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | Descriptor: | Neurogenic locus notch homolog protein 1 | | Authors: | Hosoguchi, K, Shimizu, K, Fujitani, N, Nishimura, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
2RQZ
 
 | | Structure of sugar modified epidermal growth factor-like repeat 12 of mouse Notch-1 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-L-fucopyranose, Neurogenic locus notch homolog protein 1 | | Authors: | Shimizu, K, Fujitani, N, Hosoguchi, K, Nishimura, S. | | Deposit date: | 2010-02-26 | | Release date: | 2010-10-13 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | Chemical Synthesis, Folding, and Structural Insights into O-Fucosylated Epidermal Growth Factor-like Repeat 12 of Mouse Notch-1 Receptor
J.Am.Chem.Soc., 132, 2010
|
|
2RSC
 
 | |
2V93
 
 | | EQUILLIBRIUM MIXTURE OF OPEN AND PARTIALLY-CLOSED SPECIES IN THE APO STATE OF MALTODEXTRIN-BINDING PROTEIN BY PARAMAGNETIC RELAXATION ENHANCEMENT NMR | | Descriptor: | 1-(1-HYDROXY-2,2,6,6-TETRAMETHYLPIPERIDIN-4-YL)PYRROLIDINE-2,5-DIONE, MALTOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Clore, G.M, Tang, C. | | Deposit date: | 2007-08-21 | | Release date: | 2007-11-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Open-to-Closed Transition in Apo Maltose-Binding Protein Observed by Paramagnetic NMR.
Nature, 449, 2007
|
|
2VJ3
 
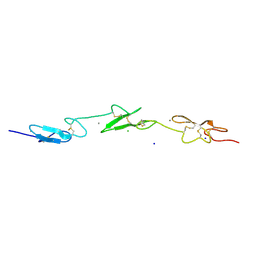 | | Human Notch-1 EGFs 11-13 | | Descriptor: | CALCIUM ION, CHLORIDE ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1, ... | | Authors: | Johnson, S, Cordle, J, Tay, J.Z, Roversi, P, Lea, S.M. | | Deposit date: | 2007-12-06 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Conserved Face of the Jagged/Serrate Dsl Domain is Involved in Notch Trans-Activation and Cis-Inhibition.
Nat.Struct.Mol.Biol., 15, 2008
|
|
4HZW
 
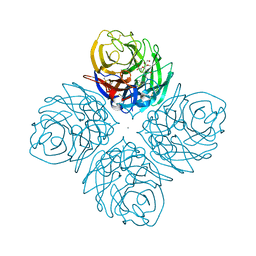 | | Crystal structure of influenza A neuraminidase N3 complexed with laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J, Vavricka, C.J, Gao, G.F. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Functional and structural analysis of influenza virus neuraminidase N3 offers further insight into the mechanisms of oseltamivir resistance.
J.Virol., 87, 2013
|
|
2VRW
 
 | |
4GLI
 
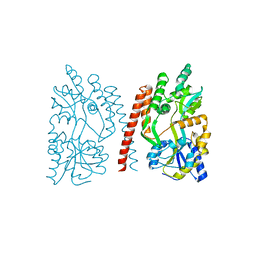 | | Crystal Structure of Human SMN YG-Dimer | | Descriptor: | Maltose-binding periplasmic protein, Survival motor neuron protein chimera | | Authors: | Martin, R.S, Perry, K, Van Duyne, G.D. | | Deposit date: | 2012-08-14 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | The survival motor neuron protein forms soluble glycine zipper oligomers.
Structure, 20, 2012
|
|
4GIZ
 
 | | Crystal structure of full-length human papillomavirus oncoprotein E6 in complex with LXXLL peptide of ubiquitin ligase E6AP at 2.55 A resolution | | Descriptor: | Maltose-binding periplasmic protein, UBIQUITIN LIGASE EA6P: chimeric protein, Protein E6, ... | | Authors: | McEwen, A.G, Zanier, K, Charbonnier, S, Poussin, P, Cura, V, Vande Pol, S, Trave, G, Cavarelli, J. | | Deposit date: | 2012-08-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins.
Science, 339, 2013
|
|
4GIF
 
 | | C-terminal coiled-coil domain of transient receptor potential channel TRPP3 (PKD2L1, Polycystin-L) | | Descriptor: | Polycystic kidney disease 2-like 1 protein | | Authors: | Yu, Y, Ulbrich, M.H, Li, M.-H, Dobbins, S, Zhang, W.K, Tong, L, Isacoff, E.Y, Yang, J. | | Deposit date: | 2012-08-08 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular mechanism of the assembly of an acid-sensing receptor ion channel complex.
Nat Commun, 3, 2012
|
|
2VGQ
 
 | | Crystal Structure of Human IPS-1 CARD | | Descriptor: | SULFATE ION, Sugar ABC transporter substrate-binding protein,Mitochondrial antiviral-signaling protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Potter, J.A, Randall, R.E, Taylor, G.L. | | Deposit date: | 2007-11-15 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Ips-1 Caspase Activation Recruitment Domain
Bmc Struct.Biol., 8, 2008
|
|
7NZM
 
 | | Cryo-EM structure of pre-dephosphorylation complex of phosphorylated eIF2alpha with trapped holophosphatase (PP1A_D64A/PPP1R15A/G-actin/DNase I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Yan, Y, Hardwick, S, Ron, D. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Higher-order phosphatase-substrate contacts terminate the integrated stress response.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4H1G
 
 | | Structure of Candida albicans Kar3 motor domain fused to maltose-binding protein | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Allingham, J.A, Duan, D, Delorme, C, Joshi, M. | | Deposit date: | 2012-09-10 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the Candida albicans Kar3 kinesin motor domain fused to maltose-binding protein.
Biochem.Biophys.Res.Commun., 428, 2012
|
|
4IL0
 
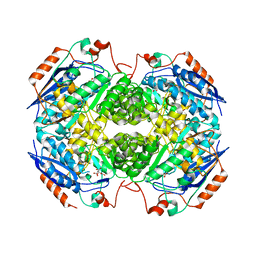 | | Crystal structure of GlucDRP from E. coli K-12 MG1655 (EFI target EFI-506058) | | Descriptor: | CITRIC ACID, GLYCEROL, Glucarate dehydratase-related protein | | Authors: | Lukk, T, Ghasempur, S, Imker, H.J, Gerlt, J.A, Nair, S.K, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-12-28 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Glucarate dehydratase and its related protein from Escherichia coli form a heterotetrameric complex.
to be published
|
|
7OJN
 
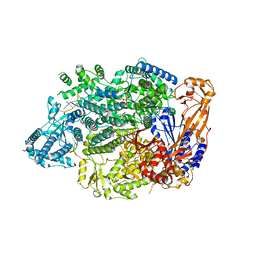 | | Lassa virus L protein in an elongation conformation [ELONGATION] | | Descriptor: | 3' RNA, 5' RNA, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, ... | | Authors: | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
7O2W
 
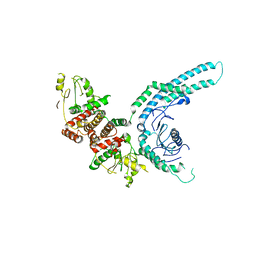 | | Structure of the C9orf72-SMCR8 complex | | Descriptor: | Guanine nucleotide exchange protein SMCR8,Guanine nucleotide exchange protein SMCR8,Maltose/maltodextrin-binding periplasmic protein, Ubiquitin-like protein SMT3,Guanine nucleotide exchange C9orf72 | | Authors: | Noerpel, J, Cavadini, S, Schenk, A.D, Graff-Meyer, A, Chao, J, Bhaskar, V. | | Deposit date: | 2021-03-31 | | Release date: | 2021-07-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structure of the human C9orf72-SMCR8 complex reveals a multivalent protein interaction architecture.
Plos Biol., 19, 2021
|
|
