1XXA
 
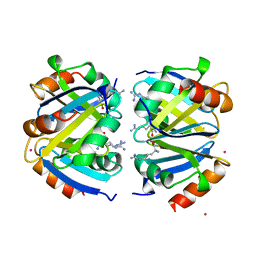 | | C-TERMINAL DOMAIN OF ESCHERICHIA COLI ARGININE REPRESSOR/ L-ARGININE COMPLEX; PB DERIVATIVE | | Descriptor: | ARGININE, ARGININE REPRESSOR, LEAD (II) ION | | Authors: | Van Duyne, G.D, Ghosh, G, Maas, W.K, Sigler, P.B. | | Deposit date: | 1995-11-03 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the oligomerization and L-arginine binding domain of the arginine repressor of Escherichia coli.
J.Mol.Biol., 256, 1996
|
|
1XXB
 
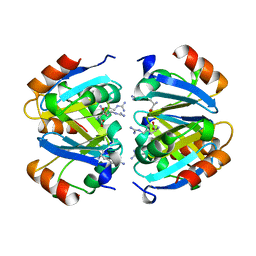 | | C-TERMINAL DOMAIN OF ESCHERICHIA COLI ARGININE REPRESSOR/ L-ARGININE COMPLEX | | Descriptor: | ARGININE, ARGININE REPRESSOR | | Authors: | Van Duyne, G.D, Ghosh, G, Maas, W.K, Sigler, P.B. | | Deposit date: | 1995-11-03 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the oligomerization and L-arginine binding domain of the arginine repressor of Escherichia coli.
J.Mol.Biol., 256, 1996
|
|
1XXC
 
 | |
1FKB
 
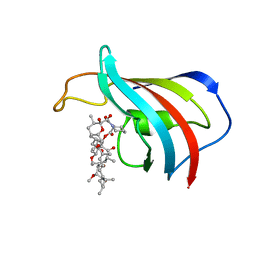 | | ATOMIC STRUCTURE OF THE RAPAMYCIN HUMAN IMMUNOPHILIN FKBP-12 COMPLEX | | Descriptor: | FK506 BINDING PROTEIN, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Van Duyne, G.D, Standaert, R.F, Schreiber, S.L, Clardy, J.C. | | Deposit date: | 1992-07-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atomic Structure of the Rapamycin Human Immunophilin Fkbp-12 Complex
J.Am.Chem.Soc., 113, 1991
|
|
5CRX
 
 | |
4RG5
 
 | | Crystal Structure of S. Pombe SMN YG-Dimer | | Descriptor: | MALONATE ION, Maltose-binding periplasmic protein, Survival Motor Neuron protein chimera, ... | | Authors: | Gupta, K, Martin, R.S, Sarachan, K.L, Sharp, B, Van Duyne, G.D. | | Deposit date: | 2014-09-29 | | Release date: | 2015-07-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Oligomeric Properties of Survival Motor NeuronGemin2 Complexes.
J.Biol.Chem., 290, 2015
|
|
4CRX
 
 | |
1B4B
 
 | | STRUCTURE OF THE OLIGOMERIZATION DOMAIN OF THE ARGININE REPRESSOR FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | ARGININE, ARGININE REPRESSOR | | Authors: | Ni, J, Sakanyan, V, Charlier, D, Glansdorff, N, Van Duyne, G.D. | | Deposit date: | 1998-12-18 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the arginine repressor from Bacillus stearothermophilus.
Nat.Struct.Biol., 6, 1999
|
|
5U96
 
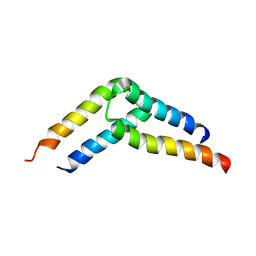 | |
5UAE
 
 | | Crystal structure of the coiled-coil domain from Listeria Innocua Phage Integrase (Trigonal Form) | | Descriptor: | CITRATE ANION, Putative integrase | | Authors: | Gupta, K, Sharp, R, Yuan, J.B, Van Duyne, G.D. | | Deposit date: | 2016-12-19 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Coiled-coil interactions mediate serine integrase directionality.
Nucleic Acids Res., 45, 2017
|
|
5UDO
 
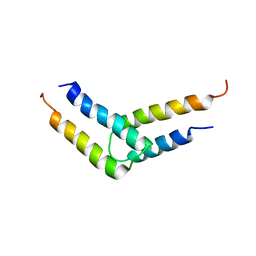 | |
6DNW
 
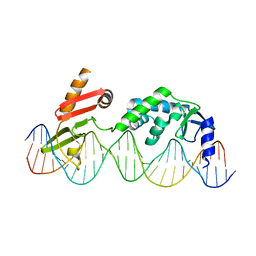 | | Sequence Requirements of the Listeria innocua prophage attP site | | Descriptor: | DNA (26-MER), Putative integrase [Bacteriophage A118], ZINC ION | | Authors: | Li, H, Sharp, R, Rutherford, K, Gupta, K, Van Duyne, G.D. | | Deposit date: | 2018-06-08 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.849 Å) | | Cite: | Serine Integrase attP Binding and Specificity.
J. Mol. Biol., 430, 2018
|
|
1B4A
 
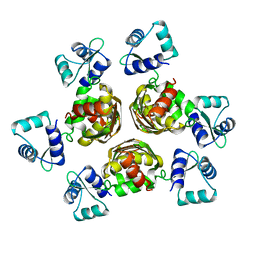 | | STRUCTURE OF THE ARGININE REPRESSOR FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | ARGININE REPRESSOR | | Authors: | Ni, J, Sakanyan, V, Charlier, D, Glansdorff, N, Van Duyne, G.D. | | Deposit date: | 1998-12-18 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the arginine repressor from Bacillus stearothermophilus.
Nat.Struct.Biol., 6, 1999
|
|
5HOT
 
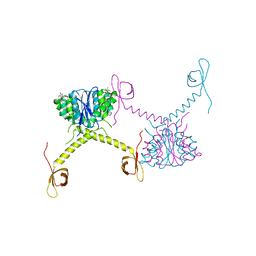 | | Structural Basis for Inhibitor-Induced Aggregation of HIV-1 Integrase | | Descriptor: | (2S)-tert-butoxy[4-(8-fluoro-5-methyl-3,4-dihydro-2H-chromen-6-yl)-2-methyl-1-oxo-1,2-dihydroisoquinolin-3-yl]ethanoic acid, Integrase | | Authors: | Gupta, K, Turkki, V, Sherrill-Mix, S, Hwang, Y, Eilers, G, Taylor, L, McDanal, C, Wang, P, Temelkoff, D, Nolte, R, Velthuisen, E, Jeffrey, J, Van Duyne, G.D, Bushman, F.D. | | Deposit date: | 2016-01-19 | | Release date: | 2016-12-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
6VRG
 
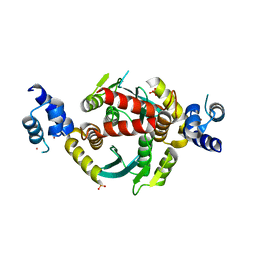 | | Structure of HIV-1 integrase with native amino-terminal sequence | | Descriptor: | Integrase, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Eilers, G, Gupta, K, Allen, A, Zhou, J, Hwang, Y, Cory, M, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2020-02-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Influence of the amino-terminal sequence on the structure and function of HIV integrase.
Retrovirology, 17, 2020
|
|
3MGV
 
 | | Cre recombinase-DNA transition state | | Descriptor: | DNA (5'-D(*CP*AP*TP*AP*TP*GP*CP*TP*AP*TP*AP*CP*GP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*CP*TP*TP*CP*GP*TP*AP*TP*AP*G)-3'), Recombinase cre, ... | | Authors: | Gibb, B.P, Gupta, K, Ghosh, K, Sharp, R, Chen, J, Van Duyne, G.D. | | Deposit date: | 2010-04-07 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Requirements for catalysis in the Cre recombinase active site.
Nucleic Acids Res., 38, 2010
|
|
3IGC
 
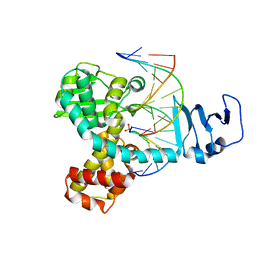 | | Smallpox virus topoisomerase-DNA transition state | | Descriptor: | 5'-D(*AP*TP*TP*CP*C)-3', 5'-D(*CP*GP*GP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*AP*CP*A)-3', 5'-D(*GP*TP*GP*TP*CP*GP*CP*CP*CP*TP*T)-3', ... | | Authors: | Perry, K, Hwang, Y, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2009-07-27 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights from the Structure of a Smallpox Virus Topoisomerase-DNA Transition State Mimic.
Structure, 18, 2010
|
|
5E6F
 
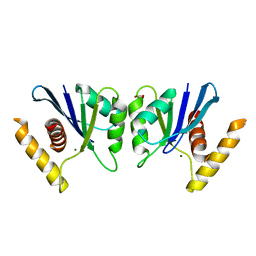 | | Canarypox virus resolvase | | Descriptor: | CNPV261 Holliday junction resolvase protein, D(-)-TARTARIC ACID, MAGNESIUM ION | | Authors: | Li, H, Hwang, Y, Perry, K, Bushman, F.D, Van Duyne, G.D. | | Deposit date: | 2015-10-09 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Metal Binding Properties of a Poxvirus Resolvase.
J.Biol.Chem., 291, 2016
|
|
8CT5
 
 | |
8CT7
 
 | | Catalytic Core Domain of HIV-1 Integrase (F185K) bound with BI-224436 | | Descriptor: | (2S)-tert-butoxy[4-(2,3-dihydropyrano[4,3,2-de]quinolin-7-yl)-2-methylquinolin-3-yl]acetic acid, 1,2-ETHANEDIOL, Integrase, ... | | Authors: | Gupta, K, Van Duyne, G.D, Eilers, G, Bushman, F.D. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of a HIV-1 IN-Allosteric inhibitor complex at 2.93 angstrom resolution: Routes to inhibitor optimization.
Plos Pathog., 19, 2023
|
|
8CTA
 
 | | Minimal 2:2 Ternary Complex between BI-224436 bound HIV-1 Integrase Catalytic Core Domain Dimer and Carboxy Terminal Domains | | Descriptor: | (2S)-tert-butoxy[4-(2,3-dihydropyrano[4,3,2-de]quinolin-7-yl)-2-methylquinolin-3-yl]acetic acid, 1,2-ETHANEDIOL, Integrase | | Authors: | Gupta, K, Van Duyne, G.D, Eilers, G, Bushman, F.D. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structure of a HIV-1 IN-Allosteric inhibitor complex at 2.93 angstrom resolution: Routes to inhibitor optimization.
Plos Pathog., 19, 2023
|
|
4GLI
 
 | | Crystal Structure of Human SMN YG-Dimer | | Descriptor: | Maltose-binding periplasmic protein, Survival motor neuron protein chimera | | Authors: | Martin, R.S, Perry, K, Van Duyne, G.D. | | Deposit date: | 2012-08-14 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | The survival motor neuron protein forms soluble glycine zipper oligomers.
Structure, 20, 2012
|
|
4KIS
 
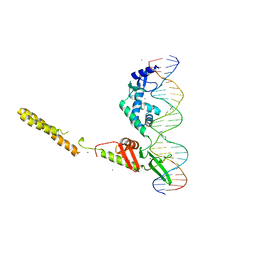 | | Crystal Structure of a LSR-DNA Complex | | Descriptor: | CALCIUM ION, DNA (26-MER), Putative integrase [Bacteriophage A118], ... | | Authors: | Rutherford, K, Yuan, P, Perry, K, Van Duyne, G.D. | | Deposit date: | 2013-05-02 | | Release date: | 2013-07-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Attachment site recognition and regulation of directionality by the serine integrases.
Nucleic Acids Res., 41, 2013
|
|
3C29
 
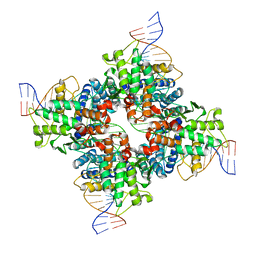 | | Cre-loxP Synaptic structure | | Descriptor: | LoxP DNA, chain C,, chain D,F, ... | | Authors: | Ghosh, K, Van Duyne, G.D. | | Deposit date: | 2008-01-24 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synapsis study in detail
To be Published
|
|
3C28
 
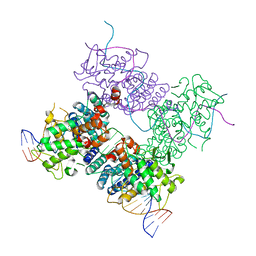 | |
