5DPE
 
 | | Thermolysin in complex with inhibitor. | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2015-09-12 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Thermodynamics of protein-ligand interactions as a reference for computational analysis: how to assess accuracy, reliability and relevance of experimental data.
J. Comput. Aided Mol. Des., 29, 2015
|
|
5DPF
 
 | | Thermolysin in complex with inhibitor. | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2015-09-12 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Thermodynamics of protein-ligand interactions as a reference for computational analysis: how to assess accuracy, reliability and relevance of experimental data.
J. Comput. Aided Mol. Des., 29, 2015
|
|
4Y37
 
 | | Endothiapepsin in complex with fragement 305 | | Descriptor: | 4-chloro-2-methylthieno[2,3-d][1,2,3]diazaborinin-1(2H)-ol, ACETATE ION, Endothiapepsin, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y3G
 
 | |
4Y3X
 
 | |
5MA7
 
 | | Structure of thermolysin in complex with inhibitor (JC306). | | Descriptor: | (2~{S})-2-[[(2~{S})-3-azanyl-2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]propanoyl]amino]-4-methyl-pentanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-11-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | How Nothing Boosts Affinity: Hydrophobic Ligand Binding to the Virtually Vacated S1' Pocket of Thermolysin.
J. Am. Chem. Soc., 139, 2017
|
|
5M69
 
 | | Thermolysin in complex with inhibitor and xenon | | Descriptor: | (2~{S})-4-methyl-2-[2-[[oxidanyl(phenylmethoxycarbonylaminomethyl)phosphoryl]amino]ethanoylamino]pentanoic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Krimmer, S.G, Cramer, J, Heine, A, Klebe, G. | | Deposit date: | 2016-10-24 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | How Nothing Boosts Affinity: Hydrophobic Ligand Binding to the Virtually Vacated S1' Pocket of Thermolysin.
J. Am. Chem. Soc., 139, 2017
|
|
8DFM
 
 | | Ectodomain of full-length wild-type KIT-SCF dimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Mast/stem cell growth factor receptor Kit, ... | | Authors: | Krimmer, S.G, Bertoletti, N, Mi, W, Schlessinger, J. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Cryo-EM analyses of KIT and oncogenic mutants reveal structural oncogenic plasticity and a target for therapeutic intervention.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8DFQ
 
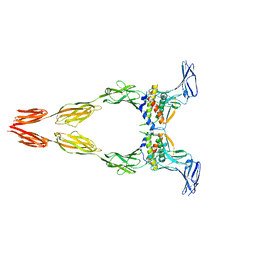 | | Ectodomain of full-length KIT(T417I,delta418-419)-SCF dimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Mast/stem cell growth factor receptor Kit, ... | | Authors: | Krimmer, S.G, Bertoletti, N, Mi, W, Schlessinger, J. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM analyses of KIT and oncogenic mutants reveal structural oncogenic plasticity and a target for therapeutic intervention.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6B1C
 
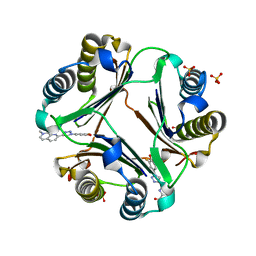 | | Macrophage Migration Inhibitory Factor in complex with a Naphthyridinone Inhibitor (4a) | | Descriptor: | 2-[1-(3-fluoro-4-hydroxyphenyl)-1H-1,2,3-triazol-4-yl]-7-methyl-1,7-naphthyridin-8(7H)-one, Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Krimmer, S.G, Robertson, M.J, Jorgensen, W.L. | | Deposit date: | 2017-09-18 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.163 Å) | | Cite: | Adding a Hydrogen Bond May Not Help: Naphthyridinone vs Quinoline Inhibitors of Macrophage Migration Inhibitory Factor.
ACS Med Chem Lett, 8, 2017
|
|
6B2C
 
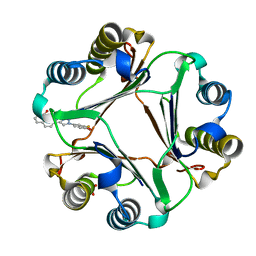 | | Macrophage Migration Inhibitory Factor in Complex with a Naphthyridinone Inhibitor (4b) | | Descriptor: | Macrophage migration inhibitory factor, SULFATE ION, {2-[1-(3-fluoro-4-hydroxyphenyl)-1H-1,2,3-triazol-4-yl]-8-oxo-1,7-naphthyridin-7(8H)-yl}acetic acid | | Authors: | Krimmer, S.G, Robertson, M.J, Jorgensen, W.L. | | Deposit date: | 2017-09-19 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Adding a Hydrogen Bond May Not Help: Naphthyridinone vs Quinoline Inhibitors of Macrophage Migration Inhibitory Factor.
ACS Med Chem Lett, 8, 2017
|
|
6B1K
 
 | | Macrophage Migration Inhibitory Factor in Complex with a Naphthyridinone Inhibitor (3a) | | Descriptor: | 2-[1-(4-hydroxyphenyl)-1H-1,2,3-triazol-4-yl]-7-methyl-1,7-naphthyridin-8(7H)-one, GLYCEROL, Macrophage migration inhibitory factor, ... | | Authors: | Krimmer, S.G, Robertson, M.J, Jorgensen, W.L. | | Deposit date: | 2017-09-18 | | Release date: | 2018-01-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Adding a Hydrogen Bond May Not Help: Naphthyridinone vs Quinoline Inhibitors of Macrophage Migration Inhibitory Factor.
ACS Med Chem Lett, 8, 2017
|
|
7SZW
 
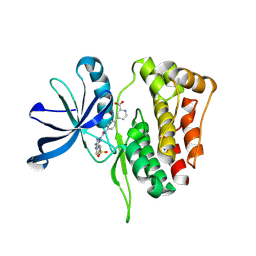 | | JAK2 JH2 in complex with JAK249 | | Descriptor: | 4-(4-{[5-amino-3-(4-sulfamoylanilino)-1H-1,2,4-triazole-1-carbonyl]amino}phenyl)pyridine-2-carboxylic acid, Tyrosine-protein kinase JAK2 | | Authors: | Krimmer, S.G, Liosi, M.E, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2021-11-29 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.90646267 Å) | | Cite: | Insights on JAK2 Modulation by Potent, Selective, and Cell-Permeable Pseudokinase-Domain Ligands.
J.Med.Chem., 65, 2022
|
|
6CB5
 
 | |
4Y3S
 
 | |
4Y47
 
 | | Endothiapepsin in complex with fragment 162 | | Descriptor: | 4-oxo-N-[(1S)-1-(pyridin-3-yl)ethyl]-4-(thiophen-2-yl)butanamide, ACETATE ION, Endothiapepsin, ... | | Authors: | Krimmer, S.G, Krug, M, Uehlein, M, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y44
 
 | | Endothiapepsin in complex with fragment 164 | | Descriptor: | 4,6-dimethyl-2-{[3-(morpholin-4-yl)propyl]amino}pyridine-3-carbonitrile, ACETATE ION, Endothiapepsin, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y3N
 
 | | Endothiapepsin in complex with fragment 273 | | Descriptor: | 2-[(1S)-2-acetyl-1,2-dihydroisoquinolin-1-yl]-N,N-dimethylacetamide, ACETATE ION, Endothiapepsin, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2015-02-10 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
4Y4E
 
 | |
6OAV
 
 | | JAK2 JH2 in complex with JAK146 | | Descriptor: | 5-amino-3-[(4-cyanophenyl)amino]-N-phenyl-1H-1,2,4-triazole-1-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Krimmer, S.G, Liosi, M.E, Puleo, D.E, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2019-03-18 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.939 Å) | | Cite: | Selective Janus Kinase 2 (JAK2) Pseudokinase Ligands with a Diaminotriazole Core.
J.Med.Chem., 63, 2020
|
|
6OBB
 
 | | JAK2 JH2 in complex with JAK170 | | Descriptor: | 5-amino-N-phenyl-3-[(4-sulfamoylphenyl)amino]-1H-1,2,4-triazole-1-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Krimmer, S.G, Liosi, M.E, Puleo, D.E, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2019-03-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Selective Janus Kinase 2 (JAK2) Pseudokinase Ligands with a Diaminotriazole Core.
J.Med.Chem., 63, 2020
|
|
6OBF
 
 | | JAK2 JH2 in complex with JAK179 | | Descriptor: | GLYCEROL, Tyrosine-protein kinase JAK2, [4-({5-amino-3-[(4-sulfamoylphenyl)amino]-1H-1,2,4-triazole-1-carbonyl}amino)phenoxy]acetic acid | | Authors: | Krimmer, S.G, Liosi, M.E, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2019-03-20 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Selective Janus Kinase 2 (JAK2) Pseudokinase Ligands with a Diaminotriazole Core.
J.Med.Chem., 63, 2020
|
|
6OCC
 
 | | JAK2 JH2 in complex with JAK190 | | Descriptor: | 2-[4-({5-amino-3-[(4-sulfamoylphenyl)amino]-1H-1,2,4-triazole-1-carbonyl}amino)phenyl]-1,3-oxazole-4-carboxylic acid, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Krimmer, S.G, Liosi, M.E, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2019-03-22 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Selective Janus Kinase 2 (JAK2) Pseudokinase Ligands with a Diaminotriazole Core.
J.Med.Chem., 63, 2020
|
|
6OBL
 
 | | JAK2 JH2 in complex with JAK168 | | Descriptor: | Tyrosine-protein kinase JAK2, [4-({5-amino-3-[(4-cyanophenyl)amino]-1H-1,2,4-triazole-1-carbonyl}amino)phenoxy]acetic acid | | Authors: | Krimmer, S.G, Liosi, M.E, Schlessinger, J, Jorgensen, W.L. | | Deposit date: | 2019-03-21 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | Selective Janus Kinase 2 (JAK2) Pseudokinase Ligands with a Diaminotriazole Core.
J.Med.Chem., 63, 2020
|
|
7MK7
 
 | | Augmentor domain of augmentor-beta | | Descriptor: | ALK and LTK ligand 1,Maltodextrin-binding protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krimmer, S.G, Reshetnyak, A.V, Puleo, D.E, Schlessinger, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42815185 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
