6EDA
 
 | | Bioreductive 4-hydroxy-3-nitro-5-ureido-benzenesulfonamides selectively target the tumor-associated carbonic anhydrase isoforms IX and XII and show hypoxia-enhanced cytotoxicity against human cancer cell lines. | | Descriptor: | 4-hydroxy-3-nitro-5-({[4-(trifluoromethyl)phenyl]carbamoyl}amino)benzene-1-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Singh, S, McKenna, R, Supuran, C.T, Nocentini, A, Lomelino, C, Lucarini, E, Bartolucci, G, Mannelli, L.D.C, Ghelardini, C, Gratteri, P. | | Deposit date: | 2018-08-09 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.879 Å) | | Cite: | 4-Hydroxy-3-nitro-5-ureido-benzenesulfonamides Selectively Target the Tumor-Associated Carbonic Anhydrase Isoforms IX and XII Showing Hypoxia-Enhanced Antiproliferative Profiles.
J. Med. Chem., 61, 2018
|
|
8HE3
 
 | |
8HE0
 
 | |
7AW2
 
 | | MerTK kinase domain with type 1.5 inhibitor from a DNA-encoded library | | Descriptor: | 5-(2'-chloro-[1,1'-biphenyl]-4-yl)-N-(imidazo[1,2-a]pyridin-6-ylmethyl)-N-methyl-1,3,4-oxadiazol-2-amine, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
5TVP
 
 | |
8I1C
 
 | |
7AW3
 
 | | MerTK kinase domain with type 1 inhibitor from a DNA-encoded library | | Descriptor: | 2-(1-((5-chloro-1H-pyrrolo[2,3-b]pyridine-3-carboxamido)methyl)-2-azabicyclo[2.1.1]hexan-2-yl)-N-methyl-4-(trifluoromethyl)thiazole-5-carboxamide, Tyrosine-protein kinase Mer | | Authors: | Schimpl, M, Nissink, J.W.M, Blackett, C, Goldberg, K, Hennessy, E.J, Hardaker, E, McCoull, W, McMurray, L, Collingwood, O, Overman, R, Pflug, A, Preston, M, Rawlins, P, Rivers, E, Smith, P, Underwood, E, Truman, C, Warwicker, J, Winter, J, Woodcock, S. | | Deposit date: | 2020-11-06 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Generating Selective Leads for Mer Kinase Inhibitors-Example of a Comprehensive Lead-Generation Strategy.
J.Med.Chem., 64, 2021
|
|
8I1G
 
 | |
9FYF
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with TR06772818 | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[5-[[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]amino]-4-fluoranyl-2-[(3~{S})-3-(methylamino)piperidin-1-yl]phenyl]propanamide | | Authors: | Kraemer, A, Ong, H.W, Yang, X, Brown, J.W, Chang, E, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-07-03 | | Release date: | 2024-09-11 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Strategic Fluorination to Achieve a Potent, Selective, Metabolically Stable, and Orally Bioavailable Inhibitor of CSNK2.
Molecules, 29, 2024
|
|
4WK4
 
 | | Metal Ion and Ligand Binding of Integrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ALA-CYS-ARG-GLY-ASP-GLY-TRP-CYS, ... | | Authors: | Xia, W, Springer, T.A. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metal ion and ligand binding of integrin alpha 5 beta 1.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5D3Y
 
 | | Crystal Structure of the P-Rex1 PH domain with Inositol-(1,3,4,5)-Tetrakisphosphate Bound | | Descriptor: | INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein | | Authors: | Cash, J.N, Tesmer, J.J.G. | | Deposit date: | 2015-08-06 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Biochemical Characterization of the Catalytic Core of the Metastatic Factor P-Rex1 and Its Regulation by PtdIns(3,4,5)P3.
Structure, 24, 2016
|
|
2R9S
 
 | | c-Jun N-terminal Kinase 3 with 3,5-Disubstituted Quinoline inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Mitogen-activated protein kinase 10, N-(tert-butyl)-4-[5-(pyridin-2-ylamino)quinolin-3-yl]benzenesulfonamide, ... | | Authors: | Habel, J. | | Deposit date: | 2007-09-13 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3,5-Disubstituted quinolines as novel c-Jun N-terminal kinase inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7WU1
 
 | | Crystal structure of phospholipase D from Moritella sp. JT01 | | Descriptor: | 1,2-ETHANEDIOL, Phospholipase D, SODIUM ION | | Authors: | Wang, Y.H, Mao, X.J, Wang, J, Wang, F.H. | | Deposit date: | 2022-02-05 | | Release date: | 2023-02-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of phospholipase D from Moritella sp. JT01
To Be Published
|
|
5HHT
 
 | |
5KH5
 
 | |
6TEN
 
 | | Crystal structure of Dot1L in complex with an inhibitor (compound 11). | | Descriptor: | 3-[(4-azanyl-6-methoxy-1,3,5-triazin-2-yl)amino]-4-[[(~{S})-[2,2-bis(fluoranyl)-1,3-benzodioxol-4-yl]-(3-chloranylpyridin-2-yl)methyl]amino]benzenesulfonamide, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Scheufler, C, Stauffer, F, Be, C, Moebitz, H. | | Deposit date: | 2019-11-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | New Potent DOT1L Inhibitors forin VivoEvaluation in Mouse.
Acs Med.Chem.Lett., 10, 2019
|
|
8K8E
 
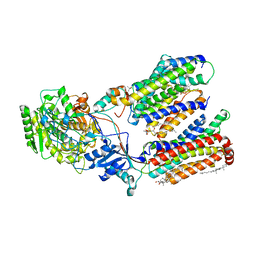 | | Human gamma-secretase in complex with a substrate mimetic | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shi, Y.G, Zhou, R, Wolfe, M.S. | | Deposit date: | 2023-07-29 | | Release date: | 2024-01-31 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Familial Alzheimer mutations stabilize synaptotoxic gamma-secretase-substrate complexes.
Cell Rep, 43, 2024
|
|
1U8K
 
 | |
8A1W
 
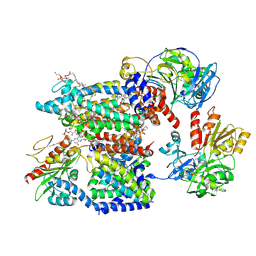 | | Sodium pumping NADH-quinone oxidoreductase with substrate Q1 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6TFZ
 
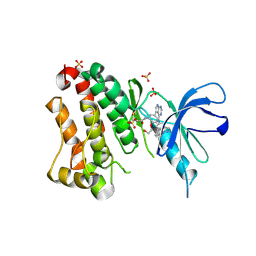 | | Crystal Structure of EGFR T790M/V948R in Complex with Covalent Pyrrolopyrimidine 19 | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, SULFATE ION, ... | | Authors: | Niggenaber, J, Mueller, M.P, Rauh, D. | | Deposit date: | 2019-11-14 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting Her2-insYVMA with Covalent Inhibitors-A Focused Compound Screening and Structure-Based Design Approach.
J.Med.Chem., 63, 2020
|
|
4Q1J
 
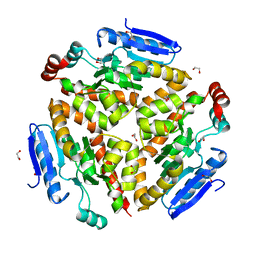 | | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-branching | | Descriptor: | 1,2-ETHANEDIOL, Polyketide biosynthesis enoyl-CoA isomerase PksI, SODIUM ION | | Authors: | Nair, A.V, Race, P.R, Till, M. | | Deposit date: | 2014-04-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-methyl branch incorporation.
Sci Rep, 10, 2020
|
|
6A3G
 
 | | Levoglucosan dehydrogenase, complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Putative dehydrogenase | | Authors: | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
5UF2
 
 | |
5K8V
 
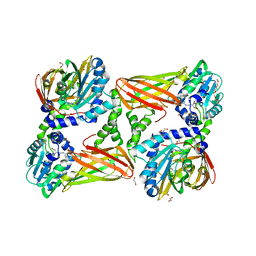 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 4 (CARM1 130-487) with CP1 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-05-31 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
7AYJ
 
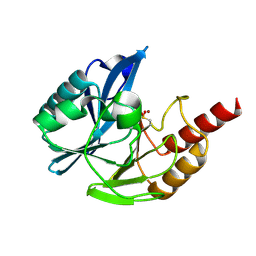 | | Metallo beta-lactamse Vim-1 with a sulfamoyl inhibitor. | | Descriptor: | 3-(4-fluorophenyl)-1-sulfamoyl-pyrrole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Farley, A.J.M, Ermolovich, I, Calvopina, K, Rabe, P, Brem, J, Schofield, C.J. | | Deposit date: | 2020-11-12 | | Release date: | 2021-05-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural Basis of Metallo-beta-lactamase Inhibition by N -Sulfamoylpyrrole-2-carboxylates.
Acs Infect Dis., 7, 2021
|
|
