1EB3
 
 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 4,7-DIOXOSEBACIC ACID COMPLEX | | Descriptor: | 4,7-DIOXOSEBACIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Stauffer, F, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2001-07-18 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Two Diacid Inhibitors
FEBS Lett., 503, 2001
|
|
5MVS
 
 | | Crystal structure of Dot1L in complex with adenosine and inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] | | Descriptor: | ADENOSINE, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-17 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5MW4
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD7 [N-(3-(((R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl)(methyl)amino)propyl)-2-(3-(2-chloro-3-(2-methylpyridin-3-yl)benzo[b]thiophen-5-yl)ureido)acetamide] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N~2~-{[2-chloro-3-(2-methylpyridin-3-yl)-1-benzothiophen-5-yl]carbamoyl}-N-(3-{methyl[(3R)-1-(5H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]amino}propyl)glycinamide, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
5MW3
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD1 [N6-(2,6-dichlorophenyl)-N6-(pent-2-yn-1-yl)quinoline-4,6-diamine] and inhibitor CPD2 [(R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine] | | Descriptor: | (3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-amine, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Be, C, Koch, E, Gaul, C, Stauffer, F, Moebitz, H, Scheufler, C. | | Deposit date: | 2017-01-18 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery of Potent, Selective, and Structurally Novel Dot1L Inhibitors by a Fragment Linking Approach.
ACS Med Chem Lett, 8, 2017
|
|
1L6S
 
 | | Crystal Structure of Porphobilinogen Synthase Complexed with the Inhibitor 4,7-Dioxosebacic Acid | | Descriptor: | 4,7-DIOXOSEBACIC ACID, MAGNESIUM ION, PORPHOBILINOGEN SYNTHASE, ... | | Authors: | Jaffe, E.K, Kervinen, J, Martins, J, Stauffer, F, Neier, R, Wlodawer, A, Zdanov, A. | | Deposit date: | 2002-03-13 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Species-Specific Inhibition of Porphobilinogen Synthase by 4-Oxosebacic Acid
J.Biol.Chem., 277, 2002
|
|
1L6Y
 
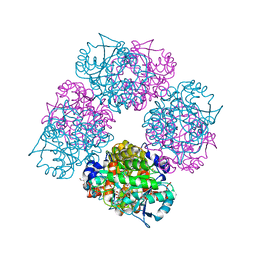 | | Crystal Structure of Porphobilinogen Synthase Complexed with the Inhibitor 4-Oxosebacic Acid | | Descriptor: | 4-OXODECANEDIOIC ACID, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Jaffe, E.K, Kervinen, J, Martins, J, Stauffer, F, Neier, R, Wlodawer, A, Zdanov, A. | | Deposit date: | 2002-03-14 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Species-Specific Inhibition of Porphobilinogen Synthase by 4-Oxosebacic Acid
J.Biol.Chem., 277, 2002
|
|
6TE6
 
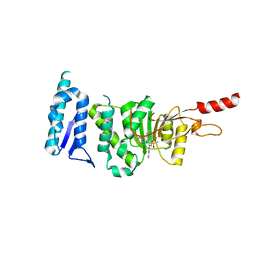 | | Crystal structure of Dot1L in complex with an inhibitor (compound 3). | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, ~{N}1-[(~{S})-(3-chlorophenyl)-pyridin-2-yl-methyl]-4-methylsulfonyl-~{N}2-pyrimidin-2-yl-benzene-1,2-diamine | | Authors: | Scheufler, C, Stauffer, F, Be, C, Moebitz, H. | | Deposit date: | 2019-11-11 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | New Potent DOT1L Inhibitors forin VivoEvaluation in Mouse.
Acs Med.Chem.Lett., 10, 2019
|
|
6TEL
 
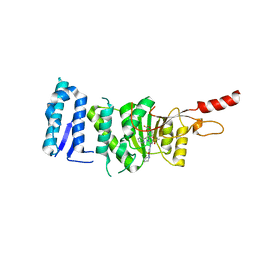 | | Crystal structure of Dot1L in complex with an inhibitor (compound 10). | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, POTASSIUM ION, ... | | Authors: | Scheufler, C, Stauffer, F, Be, C, Moebitz, H. | | Deposit date: | 2019-11-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | New Potent DOT1L Inhibitors forin VivoEvaluation in Mouse.
Acs Med.Chem.Lett., 10, 2019
|
|
6TEN
 
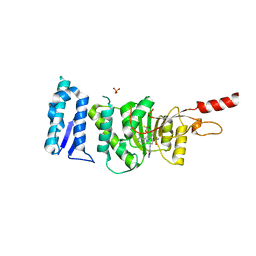 | | Crystal structure of Dot1L in complex with an inhibitor (compound 11). | | Descriptor: | 3-[(4-azanyl-6-methoxy-1,3,5-triazin-2-yl)amino]-4-[[(~{S})-[2,2-bis(fluoranyl)-1,3-benzodioxol-4-yl]-(3-chloranylpyridin-2-yl)methyl]amino]benzenesulfonamide, Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Scheufler, C, Stauffer, F, Be, C, Moebitz, H. | | Deposit date: | 2019-11-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | New Potent DOT1L Inhibitors forin VivoEvaluation in Mouse.
Acs Med.Chem.Lett., 10, 2019
|
|
1GZG
 
 | | Complex of a Mg2-dependent porphobilinogen synthase from Pseudomonas aeruginosa (mutant D139N) with 5-fluorolevulinic acid | | Descriptor: | 5-FLUOROLEVULINIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, MAGNESIUM ION, ... | | Authors: | Frere, F, Schubert, W.-D, Stauffer, F, Frankenberg, N, Neier, R, Jahn, D, Heinz, D.W. | | Deposit date: | 2002-05-21 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of porphobilinogen synthase from Pseudomonas aeruginosa in complex with 5-fluorolevulinic acid suggests a double Schiff base mechanism.
J. Mol. Biol., 320, 2002
|
|
1I8J
 
 | | CRYSTAL STRUCTURE OF PORPHOBILINOGEN SYNTHASE COMPLEXED WITH THE INHIBITOR 4,7-DIOXOSEBACIC ACID | | Descriptor: | 4,7-DIOXOSEBACIC ACID, MAGNESIUM ION, PORPHOBILINOGEN SYNTHASE, ... | | Authors: | Kervinen, J, Jaffe, E.K, Stauffer, F, Neier, R, Wlodawer, A, Zdanov, A. | | Deposit date: | 2001-03-14 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic basis for suicide inactivation of porphobilinogen synthase by 4,7-dioxosebacic acid, an inhibitor that shows dramatic species selectivity.
Biochemistry, 40, 2001
|
|
1W31
 
 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 5-HYDROXYLAEVULINIC ACID COMPLEX | | Descriptor: | 5-HYDROXYLAEVULINIC ACID, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Stauffer, F, Beaven, G.D.E, Gill, R, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2004-07-11 | | Release date: | 2005-08-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with the Inhibitor 5-Hydroxylaevulinic Acid
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5HHW
 
 | | Crystal structure of insulin receptor kinase domain in complex with cis-(R)-7-(3-(azetidin-1-ylmethyl)cyclobutyl)-5-(3-((tetrahydro-2H-pyran-2-yl)methoxy)phenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine. | | Descriptor: | 1,2-ETHANEDIOL, 7-[3-(azetidin-1-ylmethyl)cyclobutyl]-5-[3-[[(2~{R})-oxan-2-yl]methoxy]phenyl]pyrrolo[2,3-d]pyrimidin-4-amine, Insulin receptor | | Authors: | Scheufler, C, Izaac, A, Stauffer, F. | | Deposit date: | 2016-01-11 | | Release date: | 2016-04-13 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification of a 5-[3-phenyl-(2-cyclic-ether)-methylether]-4-aminopyrrolo[2,3-d]pyrimidine series of IGF-1R inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5DTR
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD5 [N-(2,6-dichlorophenyl)-4-methoxy-N-methylquinolin-6-amine] | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, N-(2,6-dichlorophenyl)-4-methoxy-N-methylquinolin-6-amine | | Authors: | Scheufler, C, Be, C, Moebitz, H, Stauffer, F. | | Deposit date: | 2015-09-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket.
Acs Med.Chem.Lett., 7, 2016
|
|
5DTQ
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD3 [(2,6-dichlorophenyl)(quinolin-6-yl)methanone] | | Descriptor: | (2,6-dichlorophenyl)(quinolin-6-yl)methanone, Histone-lysine N-methyltransferase, H3 lysine-79 specific | | Authors: | Scheufler, C, Be, C, Moebitz, H, Stauffer, F. | | Deposit date: | 2015-09-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket.
Acs Med.Chem.Lett., 7, 2016
|
|
5DTM
 
 | | Crystal structure of Dot1L in complex with inhibitor CPD1 [4-(2,6-dichlorobenzoyl)-N-methyl-1H-pyrrole-2-carboxamide] | | Descriptor: | 4-(2,6-dichlorobenzoyl)-N-methyl-1H-pyrrole-2-carboxamide, Histone-lysine N-methyltransferase, H3 lysine-79 specific | | Authors: | Scheufler, C, Be, C, Moebitz, H, Stauffer, F. | | Deposit date: | 2015-09-18 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of a Fragment-Based Screening Hit toward Potent DOT1L Inhibitors Interacting in an Induced Binding Pocket.
Acs Med.Chem.Lett., 7, 2016
|
|
