7BKP
 
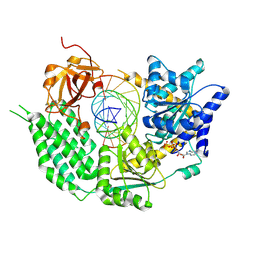 | | CryoEM structure of disease related M854K MDA5-dsRNA filament in complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Interferon-induced helicase C domain-containing protein 1, MAGNESIUM ION, ... | | 著者 | Singh, R, Herrero del Valle, A, Yu, Q, Modis, Y. | | 登録日 | 2021-01-16 | | 公開日 | 2021-11-17 | | 最終更新日 | 2021-12-01 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | MDA5 disease variant M854K prevents ATP-dependent structural discrimination of viral and cellular RNA.
Nat Commun, 12, 2021
|
|
8I5Z
 
 | |
1F3B
 
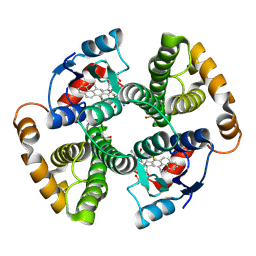 | | CRYSTAL STRUCTURE OF MGSTA1-1 IN COMPLEX WITH GLUTATHIONE CONJUGATE OF BENZO[A]PYRENE EPOXIDE | | 分子名称: | 2-AMINO-4-[1-(CARBOXYMETHYL-CARBAMOYL)-2-(9-HYDROXY-7,8-DIOXO-7,8,9,10-TETRAHYDRO-BENZO[DEF]CHRYSEN-10-YLSULFANYL)-ETHYLCARBAMOYL]-BUTYRIC ACID, GLUTATHIONE S-TRANSFERASE YA CHAIN | | 著者 | Gu, Y, Singh, S.V, Ji, X. | | 登録日 | 2000-06-01 | | 公開日 | 2000-10-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Residue R216 and catalytic efficiency of a murine class alpha glutathione S-transferase toward benzo[a]pyrene 7(R),8(S)-diol 9(S), 10(R)-epoxide.
Biochemistry, 39, 2000
|
|
1F3A
 
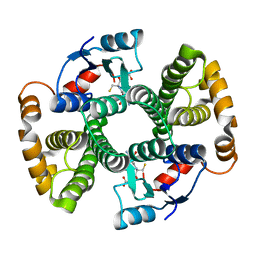 | | CRYSTAL STRUCTURE OF MGSTA1-1 IN COMPLEX WITH GSH | | 分子名称: | GLUTATHIONE, GLUTATHIONE S-TRANSFERASE YA CHAIN | | 著者 | Gu, Y, Singh, S.V, Ji, X. | | 登録日 | 2000-06-01 | | 公開日 | 2000-10-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Residue R216 and catalytic efficiency of a murine class alpha glutathione S-transferase toward benzo[a]pyrene 7(R),8(S)-diol 9(S), 10(R)-epoxide.
Biochemistry, 39, 2000
|
|
1LDM
 
 | |
2PZH
 
 | | YbgC thioesterase (Hp0496) from Helicobacter pylori | | 分子名称: | Hypothetical protein HP_0496 | | 著者 | Angelini, A, Cendron, L, Goncalves, S, Zanotti, G, Terradot, L. | | 登録日 | 2007-05-18 | | 公開日 | 2008-04-08 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and enzymatic characterization of HP0496, a YbgC thioesterase from Helicobacter pylori.
Proteins, 72, 2008
|
|
4RBM
 
 | | Porphyromonas gingivalis gingipain K (Kgp) catalytic and immunoglobulin superfamily-like domains | | 分子名称: | (3S)-3,7-diaminoheptan-2-one, ACETATE ION, AZIDE ION, ... | | 著者 | de Diego, I, Veillard, F, Sztukowska, M.N, Guevara, T, Potempa, B, Pomowski, A, Huntington, J.A, Potempa, J, Gomis-Ruth, F.X. | | 登録日 | 2014-09-12 | | 公開日 | 2014-10-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Structure and Mechanism of Cysteine Peptidase Gingipain K (Kgp), a Major Virulence Factor of Porphyromonas gingivalis in Periodontitis.
J.Biol.Chem., 289, 2014
|
|
8FLK
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP and NVS-STG2 | | 分子名称: | 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, cGAMP | | 著者 | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | 登録日 | 2022-12-21 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
8FLM
 
 | | Cryo-EM structure of STING oligomer bound to cGAMP, NVS-STG2 and C53 | | 分子名称: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, 4-({[4-(2-tert-butyl-5,5-dimethyl-1,3-dioxan-2-yl)phenyl]methyl}amino)-3-methoxybenzoic acid, Stimulator of interferon genes protein, ... | | 著者 | Li, J, Canham, S.M, Zhang, X, Bai, X, Feng, Y. | | 登録日 | 2022-12-21 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Activation of human STING by a molecular glue-like compound.
Nat.Chem.Biol., 20, 2024
|
|
8GT6
 
 | | human STING With agonist HB3089 | | 分子名称: | 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methyl-1H-furo[3,2-e]benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | 著者 | Wang, Z, Yu, X. | | 登録日 | 2022-09-07 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
8GSZ
 
 | | Structure of STING SAVI-related mutant V147L | | 分子名称: | Stimulator of interferon genes protein | | 著者 | Wang, Z, Yu, X. | | 登録日 | 2022-09-07 | | 公開日 | 2022-12-28 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.65 Å) | | 主引用文献 | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
3OQ9
 
 | | Structure of the FAS/FADD death domain assembly | | 分子名称: | Protein FADD, Tumor necrosis factor receptor superfamily member 6 | | 著者 | Kabaleeswaran, V, Wu, H. | | 登録日 | 2010-09-02 | | 公開日 | 2010-10-13 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (6.8 Å) | | 主引用文献 | The Fas-FADD death domain complex structure reveals the basis of DISC assembly and disease mutations.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7SII
 
 | | Human STING bound to both cGAMP and 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide (Compound 53) | | 分子名称: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, Stimulator of interferon genes protein, cGAMP | | 著者 | Lu, D, Shang, G, Jie, L, Lu, Y, Bai, X.C, Zhang, X. | | 登録日 | 2021-10-14 | | 公開日 | 2022-02-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.45 Å) | | 主引用文献 | Activation of STING by targeting a pocket in the transmembrane domain.
Nature, 604, 2022
|
|
2F4M
 
 | | The Mouse PNGase-HR23 Complex Reveals a Complete Remodulation of the Protein-Protein Interface Compared to its Yeast Orthologs | | 分子名称: | CHLORIDE ION, UV excision repair protein RAD23 homolog B, ZINC ION, ... | | 著者 | Zhao, G, Zhou, X, Wang, L, Kisker, C, Lennarz, W.J, Schindelin, H. | | 登録日 | 2005-11-23 | | 公開日 | 2006-03-07 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structure of the mouse peptide N-glycanase-HR23 complex suggests co-evolution of the endoplasmic reticulum-associated degradation and DNA repair pathways.
J.Biol.Chem., 281, 2006
|
|
2F4O
 
 | | The Mouse PNGase-HR23 Complex Reveals a Complete Remodulation of the Protein-Protein Interface Compared to its Yeast Orthologs | | 分子名称: | CHLORIDE ION, PHQ-VAL-ALA-ASP-CF0, XP-C repair complementing complex 58 kDa protein, ... | | 著者 | Zhao, G, Zhou, X, Wang, L, Kisker, C, Lennarz, W.J, Schindelin, H. | | 登録日 | 2005-11-23 | | 公開日 | 2006-03-07 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | Structure of the mouse peptide N-glycanase-HR23 complex suggests co-evolution of the endoplasmic reticulum-associated degradation and DNA repair pathways.
J.Biol.Chem., 281, 2006
|
|
1GLF
 
 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI GLYCEROL KINASE AND THE MUTANT A65T IN AN INACTIVE TETRAMER: CONFORMATIONAL CHANGES AND IMPLICATIONS FOR ALLOSTERIC REGULATION | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Feese, M.D, Faber, H.R, Bystrom, C.E, Pettigrew, D.W, Remington, S.J. | | 登録日 | 1998-08-30 | | 公開日 | 1998-10-16 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Glycerol kinase from Escherichia coli and an Ala65-->Thr mutant: the crystal structures reveal conformational changes with implications for allosteric regulation.
Structure, 6, 1998
|
|
6NT5
 
 | | Cryo-EM structure of full-length human STING in the apo state | | 分子名称: | Stimulator of interferon protein | | 著者 | Shang, G, Zhang, C, Chen, Z.J, Bai, X, Zhang, X. | | 登録日 | 2019-01-28 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP.
Nature, 567, 2019
|
|
6XK9
 
 | | Cereblon in complex with DDB1, CC-90009, and GSPT1 | | 分子名称: | 2-(4-chlorophenyl)-N-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1-oxo-2,3-dihydro-1H-isoindol-5-yl}methyl)-2,2-difluoroacetamide, DNA damage-binding protein 1, Eukaryotic peptide chain release factor GTP-binding subunit ERF3A, ... | | 著者 | Clayton, T.L, Tran, E.T, Zhu, J, Pagarigan, B.E, Matyskiela, M.E, Chamberlain, P.P. | | 登録日 | 2020-06-25 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | CC-90009, a novel cereblon E3 ligase modulator, targets acute myeloid leukemia blasts and leukemia stem cells.
Blood, 137, 2021
|
|
6ZME
 
 | | SARS-CoV-2 Nsp1 bound to the human CCDC124-80S-eERF1 ribosome complex | | 分子名称: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | 著者 | Thoms, M, Buschauer, R, Ameismeier, M, Denk, T, Kratzat, H, Mackens-Kiani, T, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-07-02 | | 公開日 | 2020-08-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2.
Science, 369, 2020
|
|
6XA1
 
 | |
7NIC
 
 | |
7NGA
 
 | |
7NIQ
 
 | |
7KPP
 
 | | Structure of the E102A mutant of a GNAT superfamily PA3944 acetyltransferase | | 分子名称: | 1,2-ETHANEDIOL, Acetyltransferase PA3944, COENZYME A, ... | | 著者 | Czub, M.P, Porebski, P.J, Majorek, K.A, Cymborowski, M, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-12 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
7KPS
 
 | | Structure of a GNAT superfamily PA3944 acetyltransferase in complex with AcCoA | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYL COENZYME *A, ... | | 著者 | Czub, M.P, Porebski, P.J, Cymborowski, M, Reidl, C.T, Becker, D.P, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-12 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Gcn5-Related N- Acetyltransferases (GNATs) With a Catalytic Serine Residue Can Play Ping-Pong Too.
Front Mol Biosci, 8, 2021
|
|
