4LNK
 
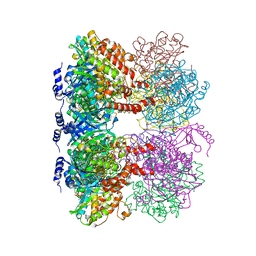 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of GS-glutamate-AMPPCP complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, GLUTAMIC ACID, Glutamine synthetase, ... | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.87 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
2L15
 
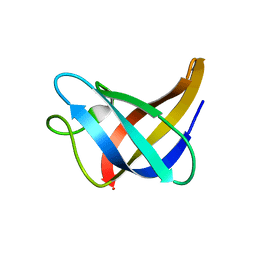 | | Solution Structure of Cold Shock Protein CspA Using Combined NMR and CS-Rosetta method | | 分子名称: | Cold shock protein CspA | | 著者 | Tang, Y, Schneider, W.M, Shen, Y, Raman, S, Inouye, M, Baker, D, Roth, M.J, Montelione, G.T. | | 登録日 | 2010-07-22 | | 公開日 | 2010-09-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Fully automated high-quality NMR structure determination of small (2)H-enriched proteins.
J Struct Funct Genomics, 11, 2010
|
|
4LL5
 
 | | Crystal Structure of Pim-1 in complex with the fluorescent compound SKF86002 | | 分子名称: | 6-(4-fluorophenyl)-5-(pyridin-4-yl)-2,3-dihydroimidazo[2,1-b][1,3]thiazole, CALCIUM ION, GLYCEROL, ... | | 著者 | Parker, L.J, Tanaka, A, Handa, N, Honda, K, Tomabechi, Y, Shirouzu, M, Yokoyama, S. | | 登録日 | 2013-07-09 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Kinase crystal identification and ATP-competitive inhibitor screening using the fluorescent ligand SKF86002.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4CFX
 
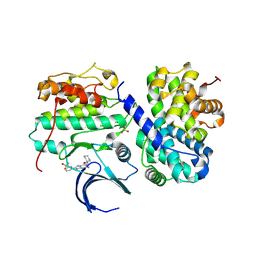 | | Structure-based design of C8-substituted O6-cyclohexylmethoxyguanine CDK1 and 2 inhibitors. | | 分子名称: | 3-[2-amino-6-(cyclohexylmethoxy)-7H-purin-8-yl]benzenesulfonamide, CYCLIN-A2, CYCLIN-DEPENDENT KINASE 2 | | 著者 | Carbain, B, Paterson, D.J, Anscombe, E, Campbell, A, Cano, C, Echalier, A, Endicott, J, Golding, B.T, Haggerty, K, Hardcastle, I.R, Jewsbury, P, Newell, D.R, Noble, M.E.M, Roche, C, Wang, L.Z, Griffin, R. | | 登録日 | 2013-11-19 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | 8-Substituted O6-Cyclohexylmethylguanine Cdk2 Inhibitors; Using Structure-Based Inhibitor Design to Optimise an Alternative Binding Mode.
J.Med.Chem., 57, 2014
|
|
4LNN
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of apo form of GS | | 分子名称: | Glutamine synthetase, MAGNESIUM ION, SULFATE ION | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-11-13 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4LNI
 
 | | B. subtilis glutamine synthetase structures reveal large active site conformational changes and basis for isoenzyme specific regulation: structure of the transition state complex | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Glutamine synthetase, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | 著者 | Schumacher, M.A, Chinnam, N, Tonthat, N, Fisher, S, Wray, L. | | 登録日 | 2013-07-11 | | 公開日 | 2013-11-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5793 Å) | | 主引用文献 | Structures of the Bacillus subtilis Glutamine Synthetase Dodecamer Reveal Large Intersubunit Catalytic Conformational Changes Linked to a Unique Feedback Inhibition Mechanism.
J.Biol.Chem., 288, 2013
|
|
4W6Z
 
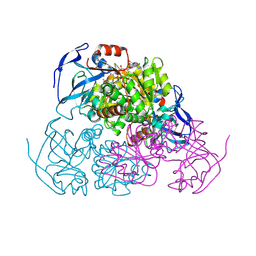 | | YEAST ALCOHOL DEHYDROGENASE I, SACCHAROMYCES CEREVISIAE FERMENTATIVE ENZYME | | 分子名称: | Alcohol dehydrogenase 1, NICOTINAMIDE-8-IODO-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | 著者 | plapp, B.v, savarimuthu, b.r, ramaswamy, s. | | 登録日 | 2014-08-21 | | 公開日 | 2014-09-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Yeast alcohol dehydrogenase structure and catalysis.
Biochemistry, 53, 2014
|
|
4X34
 
 | |
5OM7
 
 | |
4WMX
 
 | |
8AVO
 
 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (open 3:3 model). | | 分子名称: | Leptin, Leptin receptor | | 著者 | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | 登録日 | 2022-08-26 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-04-26 | | 実験手法 | ELECTRON MICROSCOPY (6.84 Å) | | 主引用文献 | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AVF
 
 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (closed 3:3 model) | | 分子名称: | Leptin, Leptin receptor | | 著者 | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | 登録日 | 2022-08-26 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-04-26 | | 実験手法 | ELECTRON MICROSCOPY (6.45 Å) | | 主引用文献 | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4WMV
 
 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 4 AT 2.4A | | 分子名称: | 3-chloro-6-fluoro-1-benzothiophene-2-carboxylic acid, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Clifton, M.C, Moulin, A. | | 登録日 | 2014-10-09 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
8AVE
 
 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (2:2 model) | | 分子名称: | Leptin, Leptin receptor | | 著者 | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | 登録日 | 2022-08-26 | | 公開日 | 2023-04-05 | | 最終更新日 | 2023-04-26 | | 実験手法 | ELECTRON MICROSCOPY (5.62 Å) | | 主引用文献 | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5OM3
 
 | | Crystal structure of Alpha1-antichymotrypsin variant DBS-I5: a MMP14-cleavable drug-binding serpin for doxycycline | | 分子名称: | (4S,4AR,5S,5AR,6R,12AS)-4-(DIMETHYLAMINO)-3,5,10,12,12A-PENTAHYDROXY-6-METHYL-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2-CARBOXAMIDE, Alpha-1-antichymotrypsin, DI(HYDROXYETHYL)ETHER | | 著者 | Schmidt, K, Muller, Y.A. | | 登録日 | 2017-07-28 | | 公開日 | 2018-05-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design of an allosterically modulated doxycycline and doxorubicin drug-binding protein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4WUY
 
 | | Crystal Structure of Protein Lysine Methyltransferase SMYD2 in complex with LLY-507, a Cell-Active, Potent and Selective Inhibitor | | 分子名称: | 5-cyano-2'-{4-[2-(3-methyl-1H-indol-1-yl)ethyl]piperazin-1-yl}-N-[3-(pyrrolidin-1-yl)propyl]biphenyl-3-carboxamide, GLYCEROL, N-lysine methyltransferase SMYD2, ... | | 著者 | Nguyen, H, Allali-Hassani, A, Antonysamy, S, Chang, S, Chen, L.H, Curtis, C, Emtage, S, Fan, L, Gheyi, T, Li, F, Liu, S, Martin, J.R, Mendel, D, Olsen, J.B, Pelletier, L, Shatseva, T, Wu, S, Zhang, F.F, Arrowsmith, C.H, Brown, P.J, Campbell, R.M, Garcia, B.A, Barsyte-Lovejoy, D, Mader, M, Vedadi, M. | | 登録日 | 2014-11-04 | | 公開日 | 2015-04-08 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | LLY-507, a Cell-active, Potent, and Selective Inhibitor of Protein-lysine Methyltransferase SMYD2.
J.Biol.Chem., 290, 2015
|
|
4WVJ
 
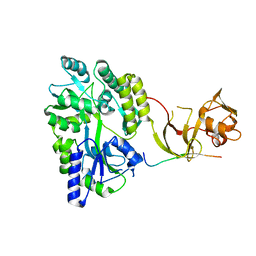 | | Crystal structure of the Type-I signal peptidase from Staphylococcus aureus (SpsB) in complex with an inhibitor peptide (pep3). | | 分子名称: | Maltose-binding periplasmic protein,Signal peptidase IB, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, inhibitor peptide (PEP3) | | 著者 | Young, P.G, Ting, Y.T, Baker, E.N. | | 登録日 | 2014-11-05 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Peptide binding to a bacterial signal peptidase visualized by peptide tethering and carrier-driven crystallization.
IUCrJ, 3, 2016
|
|
4US7
 
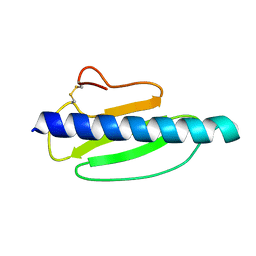 | | Sulfur SAD Phased Structure of a Type IV Pilus Protein from Shewanella oneidensis | | 分子名称: | PILD PROCESSED PROTEIN, SODIUM ION, SULFATE ION | | 著者 | Gorgel, M, Boeggild, A, Ulstrup, J.J, Mueller, U, Weiss, M, Nissen, P, Boesen, T. | | 登録日 | 2014-07-03 | | 公開日 | 2015-04-29 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | High-Resolution Structure of a Type Iv Pilin from the Metal- Reducing Bacterium Shewanella Oneidensis.
Bmc Struct.Biol., 15, 2015
|
|
4W60
 
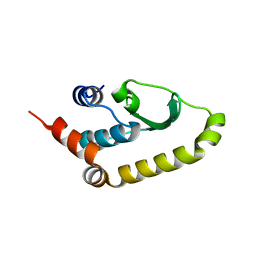 | | The structure of Vaccina virus H7 protein displays A Novel Phosphoinositide binding fold required for membrane biogenesis | | 分子名称: | Late protein H7 | | 著者 | Kolli, S, Meng, X, Wu, X, Shengjuler, D, Cameron, C.E, Xiang, Y, Deng, J. | | 登録日 | 2014-08-19 | | 公開日 | 2014-12-31 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure-function analysis of vaccinia virus h7 protein reveals a novel phosphoinositide binding fold essential for poxvirus replication.
J.Virol., 89, 2015
|
|
4WMT
 
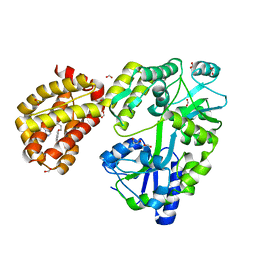 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 1 AT 2.35A | | 分子名称: | 1,2-ETHANEDIOL, 7-[2-(1H-imidazol-1-yl)-4-methylpyridin-3-yl]-3-[3-(naphthalen-1-yloxy)propyl]-1-[2-oxo-2-(piperazin-1-yl)ethyl]-1H-indole-2-carboxylic acid, FORMIC ACID, ... | | 著者 | Clifton, M.C, Dranow, D.M. | | 登録日 | 2014-10-09 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WWA
 
 | | Crystal structure of binary complex Bud32-Cgi121 | | 分子名称: | EKC/KEOPS complex subunit BUD32, EKC/KEOPS complex subunit CGI121, SULFATE ION | | 著者 | Zhang, W, van Tilbeurgh, H. | | 登録日 | 2014-11-10 | | 公開日 | 2015-03-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.953 Å) | | 主引用文献 | Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex.
Nucleic Acids Res., 43, 2015
|
|
5TBE
 
 | | Human p38alpha MAP Kinase in Complex with Dibenzosuberone Compound 2 | | 分子名称: | Mitogen-activated protein kinase 14, ~{N}-[2,4-bis(fluoranyl)-5-[[9-(2-morpholin-4-ylethylcarbamoyl)-11-oxidanylidene-5,6-dihydrodibenzo[1,2-~{d}:1',2'-~{f}][7]annulen-3-yl]amino]phenyl]thiophene-2-carboxamide | | 著者 | Buehrmann, M, Rauh, D. | | 登録日 | 2016-09-12 | | 公開日 | 2017-04-19 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Optimized Target Residence Time: Type I1/2 Inhibitors for p38 alpha MAP Kinase with Improved Binding Kinetics through Direct Interaction with the R-Spine.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2HLW
 
 | | Solution Structure of the Human Ubiquitin-conjugating Enzyme Variant Uev1a | | 分子名称: | Ubiquitin-conjugating enzyme E2 variant 1 | | 著者 | Hau, D.D, Lewis, M.J, Saltibus, L.F, Pastushok, L, Xiao, W, Spyracopoulos, L. | | 登録日 | 2006-07-10 | | 公開日 | 2006-09-05 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure and interactions of the ubiquitin-conjugating enzyme variant human uev1a: implications for enzymatic synthesis of polyubiquitin chains(,).
Biochemistry, 45, 2006
|
|
4WMU
 
 | | STRUCTURE OF MBP-MCL1 BOUND TO ligand 2 AT 1.55A | | 分子名称: | 1,2-ETHANEDIOL, 6-chloro-3-[3-(4-chloro-3,5-dimethylphenoxy)propyl]-1H-indole-2-carboxylic acid, FORMIC ACID, ... | | 著者 | Clifton, M.C, Faiman, J.W. | | 登録日 | 2014-10-09 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
4WW7
 
 | |
