9BDX
 
 | | NF-kappaB RelA homo-dimer bound to CG-centric kappaB DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*GP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*CP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), Transcription factor p65 | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDU
 
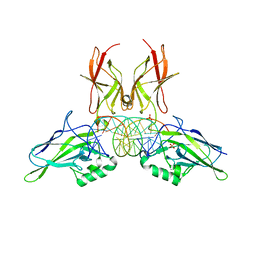 | | NF-kappaB RelA homo-dimer bound to AT-centric kappaB DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Huang, D, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDV
 
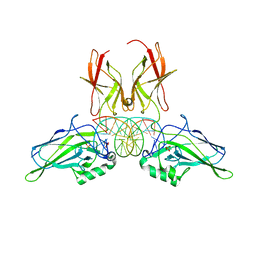 | | NF-kappaB RelA homo-dimer bound to TA-centric kappaB DNA | | Descriptor: | DNA (5'-D(*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*AP*TP*TP*CP*CP*CP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*TP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9BDW
 
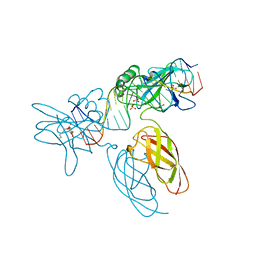 | | NF-kappaB RelA homo-dimer bound to GC-centric kappaB DNA | | Descriptor: | DNA (5'-D(P*AP*CP*TP*GP*GP*GP*AP*AP*GP*TP*TP*CP*CP*AP*GP*TP*GP*AP*T)-3'), DNA (5'-D(P*AP*TP*CP*AP*CP*TP*GP*GP*AP*AP*CP*TP*TP*CP*CP*CP*AP*GP*T)-3'), SULFATE ION, ... | | Authors: | Biswas, T, Shahabi, S, Tsodikov, O.V, Huang, D, Ghosh, G. | | Deposit date: | 2024-04-12 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Transient interactions modulate the affinity of NF-kappa B transcription factors for DNA.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8GA2
 
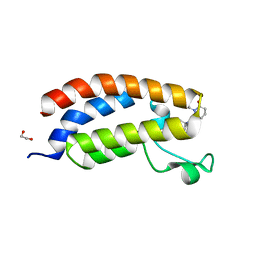 | | Bromodomain of CBP liganded with inhibitor iCBP5 | | Descriptor: | (6S)-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}-1-phenylpiperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-02-22 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8G6T
 
 | | Bromodomain of CBP liganded with inhibitor iCBP2 | | Descriptor: | (6S)-1-(3,4-dibromophenyl)-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, CREB-binding protein, NICKEL (II) ION | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FV2
 
 | | Bromodomain of CBP liganded with CCS-1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FVF
 
 | | Bromodomain of EP300 liganded with CCS-1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, Histone acetyltransferase p300, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FXE
 
 | | Bromodomain of CBP liganded with iCBP6 | | Descriptor: | (6S)-1-(3-tert-butylphenyl)-6-{(5P)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-24 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FUP
 
 | | Bromodomain of CBP liganded with BMS-536924 and CCS-1477 | | Descriptor: | (3M)-4-{[(2S)-2-(3-chlorophenyl)-2-hydroxyethyl]amino}-3-[4-methyl-6-(morpholin-4-yl)-1H-benzimidazol-2-yl]pyridin-2(1H)-one, (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-18 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of CBP and EP300 interaction with kinase inhibitors
To Be Published
|
|
8FVK
 
 | | First bromodomain of BRD4 liganded with CCS-1477 | | Descriptor: | (6S)-1-[3,4-bis(fluoranyl)phenyl]-6-[5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-(4-methoxycyclohexyl)benzimidazol-2-yl]piperidin-2-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-19 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FXA
 
 | | Bromodomain of CBP liganded with iCBP4 | | Descriptor: | (6S)-1-[3,5-bis(trifluoromethyl)phenyl]-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-24 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FXN
 
 | | Bromodomain of CBP liganded with iCBP7 | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, tert-butyl {(1R,4s)-4-[(5M)-2-[(2S)-1-(3-tert-butylphenyl)-6-oxopiperidin-2-yl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1H-benzimidazol-1-yl]cyclohexyl}carbamate | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks are sensitive to EP300/CBP bromodomain inhibition
To be published
|
|
8FXO
 
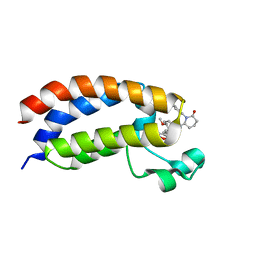 | | Bromodomain of CBP liganded with iCBP8 | | Descriptor: | (6S)-1-(2,3-dihydro-1,4-benzodioxin-6-yl)-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8I0L
 
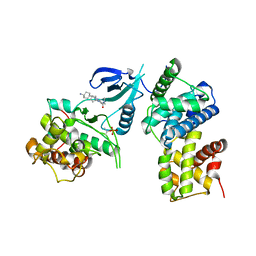 | | Structure of CDK9/cyclin T1 in complex with inhibitor | | Descriptor: | 2-[(4-azanylcyclohexyl)amino]-7-cyclopentyl-~{N},~{N}-dimethyl-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Cyclin-T1, Cyclin-dependent kinase 9 | | Authors: | Jiang, C, Ye, Y, Huang, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of CDK9/cyclin T1 in complex with inhibitor
To Be Published
|
|
8FVS
 
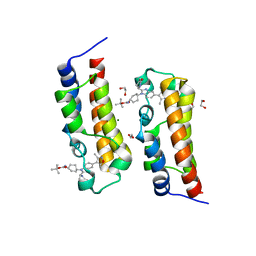 | | Bromodomain of CBP liganded with CCS1477int | | Descriptor: | 1,2-ETHANEDIOL, Histone acetyltransferase, MAGNESIUM ION, ... | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8K5R
 
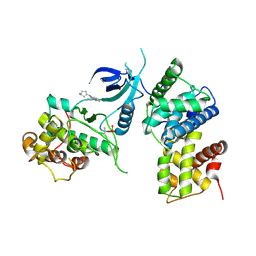 | | CDK9/cyclin T1 in complex with KB-0742 | | Descriptor: | (1S,3S)-N3-(5-pentan-3-ylpyrazolo[1,5-a]pyrimidin-7-yl)cyclopentane-1,3-diamine, Cyclin-T1, Cyclin-dependent kinase 9 | | Authors: | Zhou, M, Li, H, Gao, H, Trotter, B.W, Freeman, D. | | Deposit date: | 2023-07-24 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.751 Å) | | Cite: | Discovery of KB-0742, a Potent, Selective, Orally Bioavailable Small Molecule Inhibitor of CDK9 for MYC-Dependent Cancers.
J.Med.Chem., 66, 2023
|
|
8E1D
 
 | |
8HBM
 
 | |
8HAG
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 1 (3.2 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAH
 
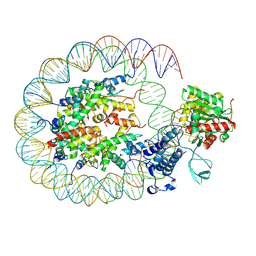 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (3.9 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAI
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 1 (4.7 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAJ
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (4.8 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAK
 
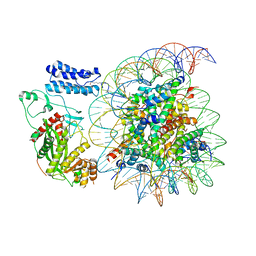 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 4 (4.5 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8CCZ
 
 | |
