6K3E
 
 | | LSD1/Co-Rest structure with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-PCPA derivative, ... | | Authors: | Wang, J. | | Deposit date: | 2019-05-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | LSD1/Co-Rest structure with an inhibitor
To Be Published
|
|
6KGK
 
 | | LSD1-CoREST-S2101 five-membered ring adduct model | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-phenylmethoxy-phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Sengoku, S, Umehara, T, Yokoyama, S. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
6KGR
 
 | | LSD1-FCPA-MPE N5 adduct model | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-[4-[5-fluoranyl-2-(trifluoromethyl)phenyl]phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Niwa, H, Sato, S, Handa, N, Umehara, T. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
6KCR
 
 | |
6KGP
 
 | | LSD1-S2157 N5 adduct model | | Descriptor: | 3-[3,5-bis(fluoranyl)-2-phenylmethoxy-phenyl]propanal, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Niwa, H, Sato, S, Umehara, T. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Development and Structural Evaluation of N-Alkylated trans-2-Phenylcyclopropylamine-Based LSD1 Inhibitors.
Chemmedchem, 15, 2020
|
|
7UWF
 
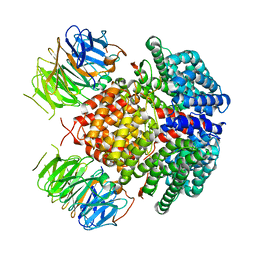 | | Human Rix1 sub-complex scaffold | | Descriptor: | Modulator of non-genomic activity of estrogen receptor, WD repeat-containing protein 18 | | Authors: | Gordon, J, Stanley, R.E. | | Deposit date: | 2022-05-03 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM reveals the architecture of the PELP1-WDR18 molecular scaffold.
Nat Commun, 13, 2022
|
|
8CQW
 
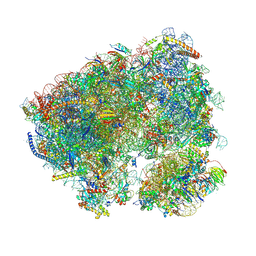 | | Crystal structure of the Candida albicans 80S ribosome in complex with Hygromycin B | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-07 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Drug-induced rotational movement of the ribosome is a key factor for read-through enhancement
To Be Published
|
|
8CRE
 
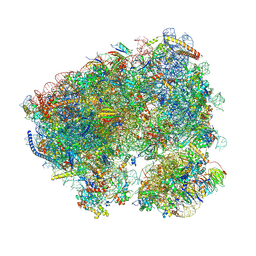 | | Crystal structure of the Candida albicans 80S ribosome in complex with geneticin G418 | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-08 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Drug-induced rotational movement of the ribosome is a key factor for read-through enhancement
To Be Published
|
|
8CQ7
 
 | | Crystal structure of phyllanthoside bound to the Candida albicans 80S ribosome | | Descriptor: | 18S, 25S, 3-O-acetyl-2-O-(3-O-acetyl-6-deoxy-beta-D-glucopyranosyl)-6-deoxy-1-O-{[(2R,2'S,3a'R,4''S,5''R,6'S,7a'S)-5''-methyl-4''-{[(2E)-3-phenylprop-2-enoyl]oxy}decahydrodispiro[oxirane-2,3'-[1]benzofuran-2',2''-pyran]-6'-yl]carbonyl}-beta-D-glucopyranose, ... | | Authors: | Kolosova, O, Zgadzay, Y, Yusupov, M. | | Deposit date: | 2023-03-03 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Drug-induced rotational movement of the ribosome is a key factor for read-through enhancement
To Be Published
|
|
6UFP
 
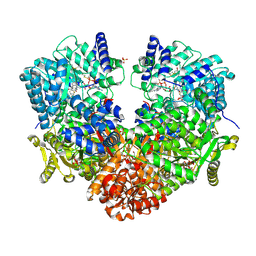 | | Structure of proline utilization A with the FAD covalently modified by L-thiazolidine-2-carboxylate and three cysteines (Cys46, Cys470, Cys638) modified to S,S-(2-HYDROXYETHYL)THIOCYSTEINE | | Descriptor: | (2S)-1,3-thiazolidine-2-carboxylic acid, Bifunctional protein PutA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Campbell, A.C, Tanner, J.J. | | Deposit date: | 2019-09-24 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Covalent Modification of the Flavin in Proline Dehydrogenase by Thiazolidine-2-Carboxylate.
Acs Chem.Biol., 15, 2020
|
|
6UGH
 
 | |
7PO5
 
 | |
7PNM
 
 | |
7PNQ
 
 | |
6V0N
 
 | | PRMT5 bound to PBM peptide from Riok1 | | Descriptor: | Methylosome protein 50, Protein arginine N-methyltransferase 5, Riok1 PBM peptide, ... | | Authors: | McMIllan, B.J, Raymond, D.D. | | Deposit date: | 2019-11-19 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Molecular basis for substrate recruitment to the PRMT5 methylosome.
Mol.Cell, 81, 2021
|
|
6UXY
 
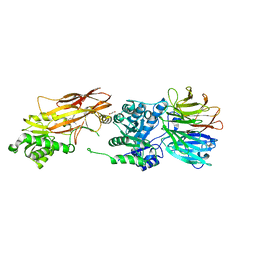 | | PRMT5:MEP50 Complexed with Allosteric Inhibitor Compound 8 | | Descriptor: | (5R)-2-amino-5-(2-cyclohexylethyl)-3-methyl-5-phenyl-3,5-dihydro-4H-imidazol-4-one, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Palte, R.L, Schneider, S.E. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Allosteric Modulation of Protein Arginine Methyltransferase 5 (PRMT5).
Acs Med.Chem.Lett., 11, 2020
|
|
5MSF
 
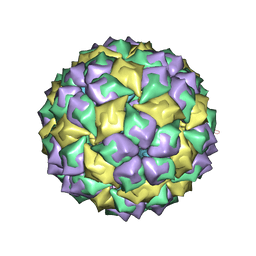 | | MS2 PROTEIN CAPSID/RNA COMPLEX | | Descriptor: | 5'-R(*CP*CP*GP*GP*AP*GP*GP*AP*UP*CP*AP*CP*CP*AP*CP*GP*GP*G)-3', MS2 PROTEIN CAPSID | | Authors: | Rowsell, S, Stonehouse, N.J, Convery, M.A, Adams, C.J, Ellington, A.D, Hirao, I, Peabody, D.S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 1998-05-15 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a series of RNA aptamers complexed to the same protein target.
Nat.Struct.Biol., 5, 1998
|
|
6B41
 
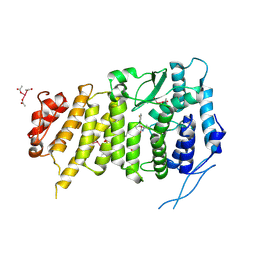 | | Menin bound to M-525 | | Descriptor: | Menin, methyl {(1S,2R)-2-[(S)-cyano[1-({1-[4-({1-[4-(dimethylamino)butanoyl]azetidin-3-yl}sulfonyl)phenyl]azetidin-3-yl}methyl)piperidin-4-yl](3-fluorophenyl)methyl]cyclopentyl}carbamate, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2017-09-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Design of the First-in-Class, Highly Potent Irreversible Inhibitor Targeting the Menin-MLL Protein-Protein Interaction.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6V0O
 
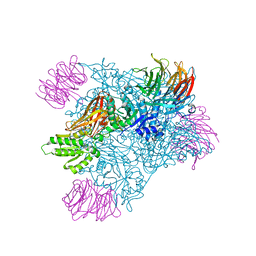 | | PRMT5 bound to the PBM peptide from pICln | | Descriptor: | ACETYL GROUP, Methylosome protein 50, PBM peptide, ... | | Authors: | McMillan, B.J, Raymond, D.D. | | Deposit date: | 2019-11-19 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Molecular basis for substrate recruitment to the PRMT5 methylosome.
Mol.Cell, 81, 2021
|
|
4ENY
 
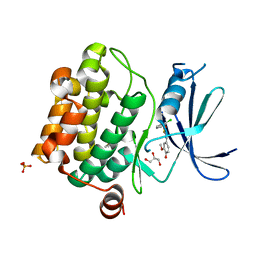 | | Crystal Structure of Pim-1 kinase in complex with (2E,5Z)-2-(2-chlorophenylimino)-5-(4-hydroxy-3-methoxybenzylidene)thiazolidin-4-one | | Descriptor: | (2Z,5Z)-2-[(2-chlorophenyl)imino]-5-(4-hydroxy-3-methoxybenzylidene)-1,3-thiazolidin-4-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-13 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Flexibility of the P-loop of Pim-1 kinase: observation of a novel conformation induced by interaction with an inhibitor
Acta Crystallogr.,Sect.F, 68, 2012
|
|
6V7Z
 
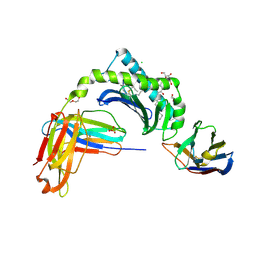 | | Human CD1d presenting alpha-Galactosylceramide in complex with VHH nanobody 1D22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d, ... | | Authors: | Shahine, A, Rossjohn, J. | | Deposit date: | 2019-12-10 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A single-domain bispecific antibody targeting CD1d and the NKT T-cell receptor induces a potent antitumor response.
Nat Cancer, 2020
|
|
8DHA
 
 | |
8DH8
 
 | |
4BZO
 
 | | Crystal structure of PIM1 in complex with a Pyrrolo-Pyrazinone inhibitor | | Descriptor: | N-[(1S)-2-AMINO-1-PHENYLETHYL]-2-[(4S)-7-(2-FLUORO-4-PYRIDINYL)-1-OXO-1,2,3,4-TETRAHYDROPYRROLO[1,2-A]PYRAZIN-4-YL]ACETAMIDE, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Casale, E, Casuscelli, F, Ardini, E, Avanzi, N, Cervi, G, D'Anello, M, Donati, D, Faiardi, D, Ferguson, R.D, Fogliatto, G, Galvani, A, Marsiglio, A, Mirizzi, D.G, Montemartini, M, Orrenius, C, Papeo, G, Piutti, C, Salom, B, Felder, E.R. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Optimization of Pyrrolo[1,2-A]Pyrazinones Leads to Novel and Selective Inhibitors of Pim Kinases.
Bioorg.Med.Chem., 21, 2013
|
|
7M3X
 
 | | Crystal Structure of the Apo Form of Human RBBP7 | | Descriptor: | Histone-binding protein RBBP7, UNKNOWN ATOM OR ION | | Authors: | Righetto, G.L, Dong, A, Li, Y, Hutchinson, A, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-19 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystal Structure of the Apo Form of Human RBBP7
To Be Published
|
|
