1MBD
 
 | |
1MBO
 
 | |
1BQ4
 
 | |
4UUI
 
 | |
5PGM
 
 | |
5OCG
 
 | | Discovery of small molecules binding to KRAS via high affinity antibody fragment competition method. | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, GTPase KRas, ... | | Authors: | Cruz-Migoni, A, Ehebauer, M.T, Phillips, S.E.V, Quevedo, C.E, Rabbitts, T.H. | | Deposit date: | 2017-06-30 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment.
Nat Commun, 9, 2018
|
|
5OCO
 
 | | Discovery of small molecules binding to KRAS via high affinity antibody fragment competition method. | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, GTPase KRas, ... | | Authors: | Cruz-Migoni, A, Ehebauer, M.T, Phillips, S.E.V, Quevedo, C.E, Rabbitts, T.H. | | Deposit date: | 2017-07-03 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment.
Nat Commun, 9, 2018
|
|
5OCT
 
 | | Discovery of small molecules binding to KRAS via high affinity antibody fragment competition method. | | Descriptor: | DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, GTPase KRas, ... | | Authors: | Cruz-Migoni, A, Ehebauer, M.T, Phillips, S.E.V, Quevedo, C.E, Rabbitts, T.H. | | Deposit date: | 2017-07-03 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment.
Nat Commun, 9, 2018
|
|
5OYM
 
 | | HIV Integrase Binding Domain of Lens Epithelium-Derived Growth Factor | | Descriptor: | PC4 and SFRS1-interacting protein | | Authors: | Rabbitts, T.H, Phillips, S.E.V, Cruz-Migoni, A, Carr, S.B, Hannon, C. | | Deposit date: | 2017-09-11 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cloning, purification and structure determination of the HIV integrase-binding domain of lens epithelium-derived growth factor.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
1BQ3
 
 | |
7MSF
 
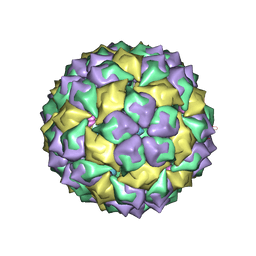 | | MS2 PROTEIN CAPSID/RNA COMPLEX | | Descriptor: | 5'-R(*UP*CP*GP*CP*CP*AP*AP*CP*AP*GP*GP*CP*GP*G)-3', MS2 PROTEIN CAPSID | | Authors: | Rowsell, S, Stonehouse, N.J, Convery, M.A, Adams, C.J, Ellington, A.D, Hirao, I, Peabody, D.S, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 1998-05-20 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of a series of RNA aptamers complexed to the same protein target.
Nat.Struct.Biol., 5, 1998
|
|
6MSF
 
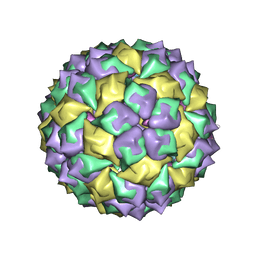 | | F6 APTAMER MS2 COAT PROTEIN COMPLEX | | Descriptor: | PROTEIN (MS2 PROTEIN CAPSID), RNA (5'-R(*CP*AP*GP*UP*CP*AP*CP*UP*GP*G)-3'), RNA (5'-R(*CP*CP*AP*CP*AP*GP*UP*CP*AP*CP*UP*GP*GP*G)-3') | | Authors: | Convery, M.A, Rowsell, S, Stonehouse, N.J, Ellington, A.D, Hirao, I, Murray, J.B, Peabody, D.S, Phillips, S.E.V, Stockley, P.G. | | Deposit date: | 1998-01-06 | | Release date: | 1998-07-08 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an RNA aptamer-protein complex at 2.8 A resolution.
Nat.Struct.Biol., 5, 1998
|
|
4V4M
 
 | | 1.45 Angstrom Structure of STNV coat protein | | Descriptor: | CALCIUM ION, Coat protein | | Authors: | Lane, S.W, Dennis, C.A, Lane, C.L, Trinh, C.H, Rizkallah, P.J, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2011-04-28 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Construction and crystal structure of recombinant STNV capsids.
J.Mol.Biol., 413, 2011
|
|
5JRD
 
 | | E. coli Hydrogenase-1 variant P508A | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Armstrong, F.A, Evans, R.M, Brooke, E.J, Islam, S.T.A. | | Deposit date: | 2016-05-06 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Importance of the Active Site "Canopy" Residues in an O2-Tolerant [NiFe]-Hydrogenase.
Biochemistry, 56, 2017
|
|
1BFV
 
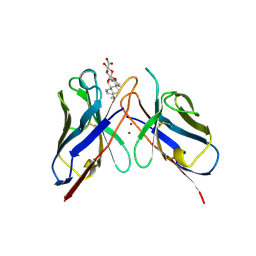 | | MONOCLONAL ANTIBODY FRAGMENT FV4155 FROM E. COLI | | Descriptor: | ESTRIOL 3-(B-D-GLUCURONIDE), FV4155, ZINC ION | | Authors: | Trinh, C.H, Phillips, S.E.V. | | Deposit date: | 1997-05-27 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibody fragment Fv4155 bound to two closely related steroid hormones: the structural basis of fine specificity.
Structure, 5, 1997
|
|
4UE3
 
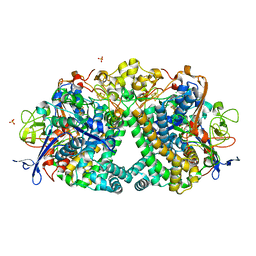 | | The Mechanism of Hydrogen Activation by NiFe-hydrogenases and the Importance of the active site Arginine | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Evans, R.M, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of Hydrogen Activation by [Nife] Hydrogenases.
Nat.Chem.Biol., 12, 2016
|
|
1T2X
 
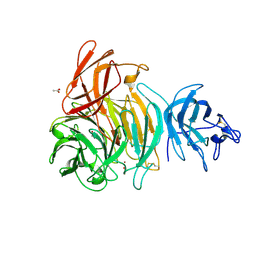 | | Glactose oxidase C383S mutant identified by directed evolution | | Descriptor: | ACETATE ION, COPPER (II) ION, Galactose Oxidase, ... | | Authors: | Wilkinson, D, Akumanyi, N, Hurtado-Guerrero, R, Dawkes, H, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and kinetic studies of a series of mutants of galactose oxidase identified by directed evolution.
Protein Eng.Des.Sel., 17, 2004
|
|
3S4G
 
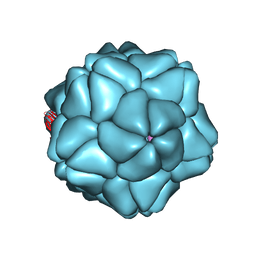 | | Low Resolution Structure of STNV complexed with RNA | | Descriptor: | Capsid protein, RNA (5'-R(P*AP*AP*A)-3'), RNA (5'-R(P*UP*UP*UP*U)-3') | | Authors: | Lane, S.W, Dennis, C.A, Lane, C.L, Trinh, C.H, Rizkallah, P.J, Stockley, P.G, Phillips, S.E.V. | | Deposit date: | 2011-05-19 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Construction and crystal structure of recombinant STNV capsids.
J.Mol.Biol., 413, 2011
|
|
3B3L
 
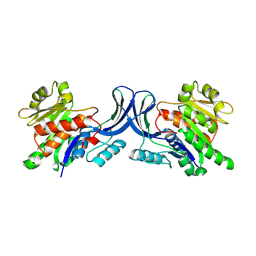 | |
1YP7
 
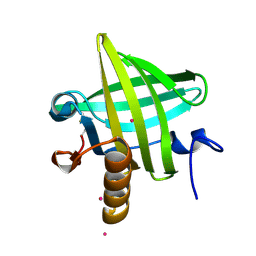 | | Van der Waals Interactions Dominate Hydrophobic Association in a Protein Binding Site Occluded From Solvent Water | | Descriptor: | CADMIUM ION, MAJOR URINARY PROTEIN 1 | | Authors: | Barratt, E, Bingham, R.J, Warner, D.J, Laughton, C.A, Phillips, S.E.V, Homans, S.W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Van der Waals Interactions Dominate Ligand-Protein Association in a Protein Binding Site Occluded from Solvent Water
J.Am.Chem.Soc., 127, 2005
|
|
5LMM
 
 | | Structure of E coli Hydrogenase Hyd-1 mutant E28Q | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Evans, R.M, Brooke, E.J, Armstrong, F.A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Kinetic consequences of re-engineering the outer shell "canopy" above the active site of a [NiFe]-hydrogenase.
To Be Published
|
|
5MVL
 
 | | Crystal structure of an A-DNA dodecamer containing 5-bromouracil | | Descriptor: | Brominated DNA dodecamer, MAGNESIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-16 | | Release date: | 2017-05-10 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.405 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVT
 
 | | Crystal structure of an A-DNA dodecamer featuring an alternating pyrimidine-purine sequence | | Descriptor: | COBALT (III) ION, DNA | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVQ
 
 | | Crystal structure of an unmodified, self-complementary dodecamer. | | Descriptor: | DNA, MAGNESIUM ION | | Authors: | Hardwick, J.S, Ptchelkine, D, Phillips, S.E.V, Brown, T. | | Deposit date: | 2017-01-17 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | 5-Formylcytosine does not change the global structure of DNA.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5MVU
 
 | |
