5GGT
 
 | | PD-L1 in complex with BMS-936559 Fab | | Descriptor: | IGK@ protein, IgG H chain, Programmed cell death 1 ligand 1 | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGR
 
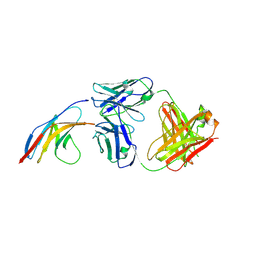 | | PD-1 in complex with nivolumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
4HZA
 
 | |
5HDO
 
 | |
5IMO
 
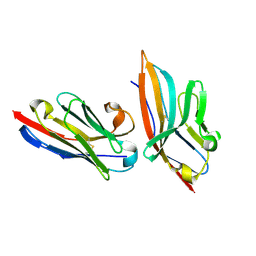 | | Nanobody targeting mouse Vsig4 in Spacegroup P3221 | | Descriptor: | Nanobody, Protein Vsig4 | | Authors: | Wen, Y, Zheng, F. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
5IR3
 
 | |
4K07
 
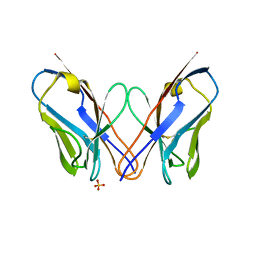 | | Crystal structure of the amyloid-forming immunoglobulin AL-103 cis-proline 95 mutant | | Descriptor: | Amyloidogenic immunoglobulin light chain protein AL-103, SULFATE ION | | Authors: | Thompson, J.R, Berkholz, D.S, Mahlum, E.W, Ramirez-Alvarado, M. | | Deposit date: | 2013-04-03 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Kinetic control in protein folding for light chain amyloidosis and the differential effects of somatic mutations.
J.Mol.Biol., 426, 2014
|
|
5IMM
 
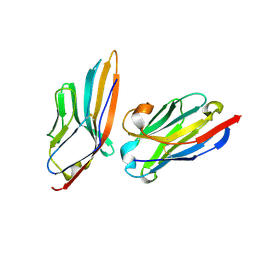 | | Nanobody targeting mouse Vsig4 in Spacegroup P212121 | | Descriptor: | Nanobody, Protein Vsig4 | | Authors: | Wen, Y, Zheng, F. | | Deposit date: | 2016-03-06 | | Release date: | 2017-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural evaluation of a nanobody targeting complement receptor Vsig4 and its cross reactivity
Immunobiology, 222, 2017
|
|
4KKN
 
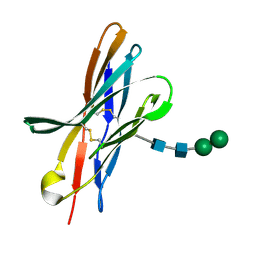 | | Crystal structure of bovine CTLA-4, PSI-NYSGRC-012704 | | Descriptor: | Cytotoxic T-lymphocyte associated protein 4, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Bonanno, J, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Scott Glenn, A, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-05-06 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structure of bovine CTLA-4, PSI-NYSGRC-012704
to be published
|
|
4JJH
 
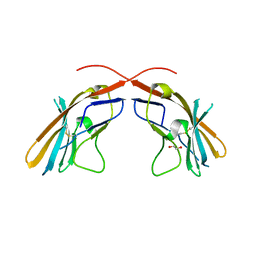 | | Crystal structure of the D1 domain from human Nectin-4 extracellular fragment [PSI-NYSGRC-005624] | | Descriptor: | 1,2-ETHANEDIOL, Poliovirus receptor-related protein 4 | | Authors: | Kumar, P.R, Bonanno, J, Ahmed, M, Banu, R, Bhosle, R, Calarese, D, Celikigil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-03-07 | | Release date: | 2013-03-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the D1 domain of human Nectin-4 extracellular fragment [NYSGRC-005624]
to be published
|
|
4K55
 
 | |
5J8O
 
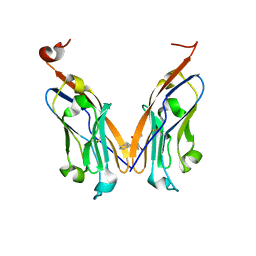 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | (2R)-1-({3-bromo-4-[(2-methyl[1,1'-biphenyl]-3-yl)methoxy]phenyl}methyl)piperidine-2-carboxylic acid, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Guzik, K, Zieba, B.J, Musielak, B, Doemling, P, Dubin, G, Holak, T.A. | | Deposit date: | 2016-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1).
Oncotarget, 7, 2016
|
|
5JDS
 
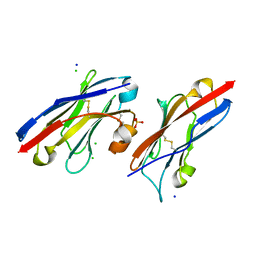 | |
5J89
 
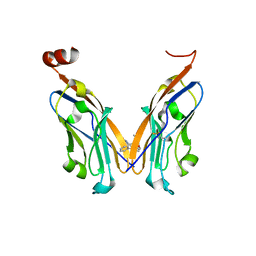 | | Structure of human Programmed cell death 1 ligand 1 (PD-L1) with low molecular mass inhibitor | | Descriptor: | 1,2-ETHANEDIOL, N-{2-[({2-methoxy-6-[(2-methyl[1,1'-biphenyl]-3-yl)methoxy]pyridin-3-yl}methyl)amino]ethyl}acetamide, Programmed cell death 1 ligand 1 | | Authors: | Zak, K.M, Grudnik, P, Guzik, K, Zieba, B.J, Musielak, B, Doemling, P, Dubin, G, Holak, T.A. | | Deposit date: | 2016-04-07 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1).
Oncotarget, 7, 2016
|
|
4JGJ
 
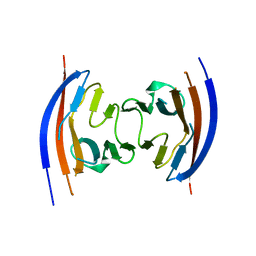 | | Crystal structure of the Ig-like D1 domain from mouse Carcinoembryogenic antigen-related cell adhesion molecule 15 (CEACAM15) [PSI-NYSGRC-005691] | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Carcinoembryonic antigen-related cell adhesion molecule 15, Unknown peptide | | Authors: | Kumar, P.R, Bonanno, J, Ahmed, M, Banu, R, Bhosle, R, Calarese, D, Celikigil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-03-01 | | Release date: | 2013-03-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6508 Å) | | Cite: | Crystal structure of the Ig-like D1 domain of CEACAM15 from Mus musculus [NYSGRC-005691]
to be published
|
|
4JKW
 
 | |
4L1H
 
 | |
4L1D
 
 | | Voltage-gated sodium channel beta3 subunit Ig domain | | Descriptor: | Sodium channel subunit beta-3 | | Authors: | Namadurai, S, Weimhofer, M, Rajappa, R, Stott, K, Klingauf, J, Chirgadze, D.Y, Jackson, A.P. | | Deposit date: | 2013-06-03 | | Release date: | 2014-03-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure and Molecular Imaging of the Nav Channel beta 3 Subunit Indicates a Trimeric Assembly.
J.Biol.Chem., 289, 2014
|
|
4DFH
 
 | | Crystal structure of cell adhesion molecule nectin-2/CD112 variable domain | | Descriptor: | Poliovirus receptor-related protein 2 | | Authors: | Liu, J, Qian, X, Chen, Z, Xu, X, Gao, F, Zhang, S, Zhang, R, Qi, J, Gao, G.F, Yan, J. | | Deposit date: | 2012-01-23 | | Release date: | 2012-06-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Cell Adhesion Molecule Nectin-2/CD112 and Its Binding to Immune Receptor DNAM-1/CD226
J.Immunol., 188, 2012
|
|
4GOS
 
 | | Crystal structure of human B7-H4 IgV-like domain | | Descriptor: | V-set domain-containing T-cell activation inhibitor 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Vigdorovich, V, Ramagopal, U, Bhosle, R, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-08-20 | | Release date: | 2012-09-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and cancer immunotherapy of the B7 family member B7x.
Cell Rep, 9, 2014
|
|
4H5S
 
 | | Complex structure of Necl-2 and CRTAM | | Descriptor: | Cell adhesion molecule 1, Cytotoxic and regulatory T-cell molecule | | Authors: | Zhang, S, Lu, G, Qi, J, Li, Y, Zhang, Z, Zhang, B, Yan, J, Gao, G.F. | | Deposit date: | 2012-09-18 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Competition of cell adhesion and immune recognition: insights into the interaction between CRTAM and nectin-like 2.
Structure, 21, 2013
|
|
5E5M
 
 | | Crystal structure of mouse CTLA-4 in complex with nanobody | | Descriptor: | CTLA-4 nanobody, Cytotoxic T-lymphocyte protein 4, GLYCEROL | | Authors: | Fedorov, A.A, Fedorov, E.V, Samanta, D, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-08 | | Release date: | 2016-10-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Crystal structure of mouse CTLA-4 in complex with nanobody
To Be Published
|
|
5DZL
 
 | | Crystal structure of the protein human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1 | | Authors: | Huang, Y.H, Russell, A, Gandhi, A.K, Kondo, Y, Chen, Q, Petsko, G.A, Blumberg, R.S. | | Deposit date: | 2015-09-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.4006 Å) | | Cite: | CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.
Nature, 517, 2015
|
|
5E56
 
 | | Crystal structure of mouse CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, SODIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, SAMANTA, D, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2015-10-07 | | Release date: | 2015-10-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.504 Å) | | Cite: | Crystal structure of mouse CTLA-4
To Be Published
|
|
5EB9
 
 | | Crystal Structure Of Chicken CD8aa Homodimer | | Descriptor: | CD8 alpha chain | | Authors: | Liu, Y.J, Qi, J.X, Xia, C. | | Deposit date: | 2015-10-18 | | Release date: | 2016-09-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | The structural basis of chicken, swine and bovine CD8 alpha alpha dimers provides insight into the co-evolution with MHC I in endotherm species.
Sci Rep, 6, 2016
|
|
