7PVC
 
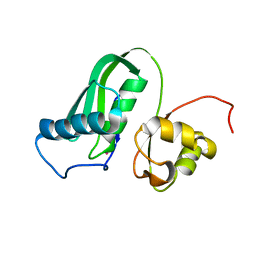 | | The structure of Kbp.K from E. coli with potassium bound. | | Descriptor: | POTASSIUM ION, Potassium binding protein Kbp | | Authors: | Smith, B.O. | | Deposit date: | 2021-10-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-11 | | Method: | SOLUTION NMR | | Cite: | Tuning the Sensitivity of Genetically Encoded Fluorescent Potassium Indicators through Structure-Guided and Genome Mining Strategies.
ACS Sens, 7, 2022
|
|
2B8H
 
 | | A/NWS/whale/Maine/1/84 (H1N9) reassortant influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Smith, B.J, Platis, D, Cox, M.M.J, Huyton, T, Joosten, R.P, McKimm-Breschkin, J.L, Zhang, J.-G, Luo, C.S, Lou, M.-Z, Garrett, T.P.J, Labrou, N.E. | | Deposit date: | 2005-10-07 | | Release date: | 2006-09-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a calcium-deficient form of influenza virus neuraminidase: implications for substrate binding.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5AGW
 
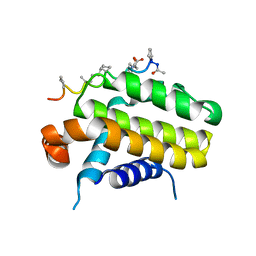 | | Bcl-2 alpha beta-1 complex | | Descriptor: | APOPTOSIS REGULATOR BCL-2, BCL-2-LIKE PROTEIN 1, BCL-2-LIKE PROTEIN 11 | | Authors: | Smith, B.J, F Lee, E, Checco, J.W, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | Alpha Beta Peptide Foldamers Targeting Intracellular Protein-Protein Interactions with Activity on Living Cells
J.Am.Chem.Soc., 137, 2015
|
|
1L7H
 
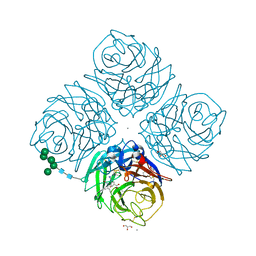 | | Crystal structure of R292K mutant influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
1L7F
 
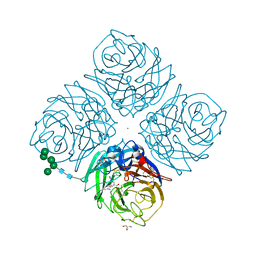 | | Crystal structure of influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
5AGX
 
 | | Bcl-2 alpha beta-1 LINEAR complex | | Descriptor: | APOPTOSIS REGULATOR BCL-2, BCL-2-LIKE PROTEIN 1, BCL-2-LIKE PROTEIN 11 | | Authors: | Smith, B.J, F Lee, E, Checco, J.W, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2015-02-04 | | Release date: | 2015-09-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Alpha Beta Peptide Foldamers Targeting Intracellular Protein-Protein Interactions with Activity on Living Cells
J.Am.Chem.Soc., 137, 2015
|
|
1TPG
 
 | | F1-G MODULE PAIR RESIDUES 1-91 (C83S) OF TISSUE-TYPE PLASMINOGEN ACTIVATOR (T-PA) (NMR, 298K, PH2.95, REPRESENTATIVE STRUCTURE) | | Descriptor: | T-PLASMINOGEN ACTIVATOR F1-G | | Authors: | Smith, B.O, Downing, A.K, Driscoll, P.C, Dudgeon, T.J, Campbell, I.D. | | Deposit date: | 1995-06-14 | | Release date: | 1995-09-15 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | The solution structure and backbone dynamics of the fibronectin type I and epidermal growth factor-like pair of modules of tissue-type plasminogen activator.
Structure, 3, 1995
|
|
5DG5
 
 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF THE HEPATOCYTE GROWTH FACTOR RECEPTOR C-MET IN COMPLEX WITH ALTIRATINIB ANALOG DP-4157 | | Descriptor: | Hepatocyte growth factor receptor, N-(2,5-difluoro-4-{[2-(1-methyl-1H-pyrazol-4-yl)pyridin-4-yl]oxy}phenyl)-N'-(4-fluorophenyl)cyclopropane-1,1-dicarboxam ide | | Authors: | Smith, B.D, Kaufman, M.D, Leary, C.B, Turner, B.A, Wise, S.A, Ahn, Y.M, Booth, R.J, Caldwell, T.M, Ensinger, C.L, Hood, M.M, Lu, W.-P, Patt, T.W, Patt, W.C, Rutkoski, T.J, Samarakoon, T, Telikepalli, H, Vogeti, L, Vogeti, S, Yates, K.M, Chun, L, Stewart, L.J, Clare, M, Flynn, D.L. | | Deposit date: | 2015-08-27 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Altiratinib Inhibits Tumor Growth, Invasion, Angiogenesis, and Microenvironment-Mediated Drug Resistance via Balanced Inhibition of MET, TIE2, and VEGFR2.
Mol.Cancer Ther., 14, 2015
|
|
1F8B
 
 | | Native Influenza Virus Neuraminidase in Complex with NEU5AC2EN | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
1F8C
 
 | | Native Influenza Neuraminidase in Complex with 4-amino-2-deoxy-2,3-dehydro-N-neuraminic Acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-AMINO-2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
1F8D
 
 | | Native Influenza Neuraminidase in Complex with 9-amino-2-deoxy-2,3-dehydro-N-neuraminic Acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 9-AMINO-2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
4BPK
 
 | | Bcl-xL bound to alpha beta Puma BH3 peptide 5 | | Descriptor: | 1,2-ETHANEDIOL, ALPHA BETA BH3-PEPTIDE, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Smith, B.J, Lee, E.F, Checco, J.W, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2013-05-27 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Structure-Guided Rational Design of Alpha/Beta-Peptide Foldamers with High Affinity for Bcl-2 Family Prosurvival Proteins.
Chembiochem, 14, 2013
|
|
1F8E
 
 | | Native Influenza Neuraminidase in Complex with 4,9-diamino-2-deoxy-2,3-dehydro-N-acetyl-neuraminic Acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,9-AMINO-2,4-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, ... | | Authors: | Smith, B.J, Colman, P.M, Von Itzstein, M, Danylec, B, Varghese, J.N. | | Deposit date: | 2000-06-30 | | Release date: | 2001-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Analysis of inhibitor binding in influenza virus neuraminidase.
Protein Sci., 10, 2001
|
|
3CH3
 
 | | Crystal Structure Analysis of SERA5E from plasmodium falciparum | | Descriptor: | DIHYDROGENPHOSPHATE ION, POTASSIUM ION, Serine-repeat antigen protein | | Authors: | Smith, B.J, Malby, R.L, Colman, P.M, Clarke, O.B. | | Deposit date: | 2008-03-07 | | Release date: | 2008-03-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Insights into the Protease-like Antigen Plasmodium falciparum SERA5 and Its Noncanonical Active-Site Serine
J.Mol.Biol., 392, 2009
|
|
2I54
 
 | | Phosphomannomutase from Leishmania mexicana | | Descriptor: | CHLORIDE ION, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Smith, B.J. | | Deposit date: | 2006-08-24 | | Release date: | 2007-04-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Leishmania mexicana phosphomannomutase highlights similarities with human isoforms
J.Mol.Biol., 363, 2006
|
|
1L7G
 
 | | Crystal structure of E119G mutant influenza virus neuraminidase in complex with BCX-1812 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, ... | | Authors: | Smith, B.J, McKimm-Breshkin, J.L, McDonald, M, Fernley, R.T, Varghese, J.N, Colman, P.M. | | Deposit date: | 2002-03-15 | | Release date: | 2002-05-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies of the resistance of influenza virus neuramindase to inhibitors.
J.Med.Chem., 45, 2002
|
|
3CH2
 
 | | Crystal Structure Analysis of SERA5E from plasmodium falciparum | | Descriptor: | CALCIUM ION, Serine-repeat antigen protein | | Authors: | Smith, B.J, Malby, R.L, Colman, P.M, Clarke, O.B. | | Deposit date: | 2008-03-07 | | Release date: | 2008-03-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Protease-like Antigen Plasmodium falciparum SERA5 and Its Noncanonical Active-Site Serine
J.Mol.Biol., 392, 2009
|
|
2I55
 
 | |
2WBF
 
 | | Crystal Structure Analysis of SERA5E from plasmodium falciparum with loop 690-700 ordered | | Descriptor: | CALCIUM ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Smith, B.J, Malby, R.L, Colman, P.M, Clarke, O.B. | | Deposit date: | 2009-02-27 | | Release date: | 2009-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the protease-like antigen Plasmodium falciparum SERA5 and its noncanonical active-site serine.
J. Mol. Biol., 392, 2009
|
|
2C34
 
 | | Leishmania mexicana ICP | | Descriptor: | INHIBITOR OF CYSTEINE PEPTIDASES | | Authors: | Smith, B.O, Picken, N.C, Bromek, K, Westrop, G.D, Mottram, J.C, Coombs, G.H. | | Deposit date: | 2005-10-04 | | Release date: | 2005-12-21 | | Last modified: | 2021-06-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of Leishmania Mexicana Icp Provides Evidence for Convergent Evolution of Cysteine Peptidase Inhibitors.
J.Biol.Chem., 281, 2006
|
|
1GKG
 
 | | Structure Determination and Rational Mutagenesis reveal binding surface of immune adherence receptor, CR1 (CD35) | | Descriptor: | COMPLEMENT RECEPTOR TYPE 1 | | Authors: | Smith, B.O, Mallin, R.L, Krych-Goldberg, M, Wang, X, Hauhart, R.E, Bromek, K, Uhrin, D, Atkinson, J.P, Barlow, P.N. | | Deposit date: | 2001-08-14 | | Release date: | 2002-04-18 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the C3B Binding Site of Cr1 (Cd35), the Immune Adherence Receptor
Cell(Cambridge,Mass.), 108, 2002
|
|
2XV9
 
 | | The solution structure of ABA-1A saturated with oleic acid | | Descriptor: | ABA-1A1 REPEAT UNIT | | Authors: | Smith, B.O, Kennedy, M.W, Cooper, A, Meenan, N.A.G, Bromek, K. | | Deposit date: | 2010-10-25 | | Release date: | 2011-08-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a repeated unit of the ABA-1 nematode polyprotein allergen of Ascaris reveals a novel fold and two discrete lipid-binding sites.
PLoS Negl Trop Dis, 5, 2011
|
|
2YQ6
 
 | | Structure of Bcl-xL bound to BimSAHB | | Descriptor: | BCL-2-LIKE PROTEIN 1, BIM BETA 5, GLYCEROL | | Authors: | Smith, B.J, Czabotar, P.E. | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Stabilizing the Pro-Apoptotic Bimbh3 Helix (Bimsahb) Does not Necessarily Enhance Affinity or Biological Activity.
Acs Chem.Biol., 8, 2013
|
|
4BPJ
 
 | | Mcl-1 bound to alpha beta Puma BH3 peptide 3 | | Descriptor: | ALPHA BETA BH3-PEPTIDE, CHLORIDE ION, FUSION PROTEIN CONSISTING OF INDUCED MYELOID LEUKEMIA CELL DIFFERENTIATION PROTEIN MCL-1 HOMOLOG, ... | | Authors: | Smith, B.J, Lee, E.F, Checco, J.W, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2013-05-27 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structure-Guided Rational Design of Alpha/Beta-Peptide Foldamers with High Affinity for Bcl-2 Family Prosurvival Proteins.
Chembiochem, 14, 2013
|
|
4BPI
 
 | | Mcl-1 bound to alpha beta Puma BH3 peptide 2 | | Descriptor: | ALPHA BETA BH3PEPTIDE, CADMIUM ION, FUSION PROTEIN CONSISTING OF INDUCED MYELOID LEUKEMIA CELL DIFFERENTIATION PROTEIN MCL-1 HOMOLOG | | Authors: | Smith, B.J, Lee, E.F, Checco, J.W, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2013-05-27 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Structure-Guided Rational Design of Alpha/Beta-Peptide Foldamers with High Affinity for Bcl-2 Family Prosurvival Proteins.
Chembiochem, 14, 2013
|
|
