7T8T
 
 | | CryoEM structure of PLCg1 | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma, CALCIUM ION | | Authors: | Endo-Streeter, S, Sondek, J. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | CryoEM structure of PLCg1
To Be Published
|
|
4AE9
 
 | | Structure and function of the Human Sperm-Specific Isoform of Protein Kinase A (PKA) Catalytic Subunit C alpha 2 | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA | | Authors: | Hereng, T.H, Backe, P.H, Kahmann, J, Scheich, C, Bjoras, M, Skalhegg, B.S, Rosendal, K.R. | | Deposit date: | 2012-01-09 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of the Human Sperm-Specific Isoform of Protein Kinase a (Pka) Catalytic Subunit Calpha2
J.Struct.Biol., 178, 2012
|
|
5NSD
 
 | | Co-crystal structure of NAMPT dimer with KPT-9274 | | Descriptor: | (~{E})-3-(6-azanylpyridin-3-yl)-~{N}-[[5-[4-[4,4-bis(fluoranyl)piperidin-1-yl]carbonylphenyl]-7-(4-fluorophenyl)-1-benzofuran-2-yl]methyl]prop-2-enamide, GLYCEROL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Neggers, J.E, Kwanten, B, Dierckx, T, Noguchi, H, Voet, A, Vercruysse, T, Baloglu, E, Senapedis, W, Jacquemyn, M, Daelemans, D. | | Deposit date: | 2017-04-26 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Target identification of small molecules using large-scale CRISPR-Cas mutagenesis scanning of essential genes.
Nat Commun, 9, 2018
|
|
1ZBF
 
 | | Crystal structure of B. halodurans RNase H catalytic domain mutant D132N | | Descriptor: | SULFATE ION, ribonuclease H-related protein | | Authors: | Nowotny, M, Gaidamakov, S.A, Crouch, R.J, Yang, W. | | Deposit date: | 2005-04-08 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis.
Cell(Cambridge,Mass.), 121, 2005
|
|
5O2R
 
 | | Cryo-EM structure of the proline-rich antimicrobial peptide Api137 bound to the terminating ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Graf, M, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2017-05-22 | | Release date: | 2017-07-26 | | Last modified: | 2019-02-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An antimicrobial peptide that inhibits translation by trapping release factors on the ribosome.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3IFC
 
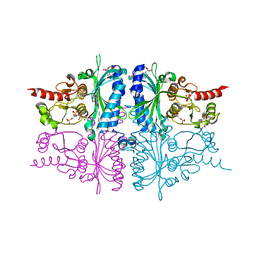 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in complex with AMP and alpha fructose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2, ... | | Authors: | Kolodziejczyk, R, Zarzycki, M, Jaskolski, M, Dzugaj, A. | | Deposit date: | 2009-07-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of E69Q mutant of human muscle fructose-1,6-bisphosphatase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5NXN
 
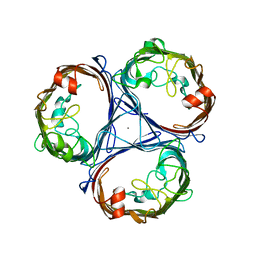 | |
5ERP
 
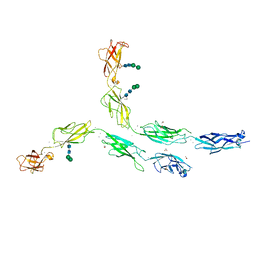 | | Crystal structure of human Desmocollin-2 ectodomain fragment EC2-5 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2015-11-14 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of adhesive binding by desmocollins and desmogleins.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7T8F
 
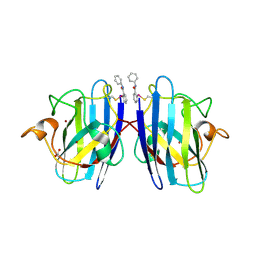 | |
5I4T
 
 | |
2CM6
 
 | | crystal structure of the C2B domain of rabphilin3A | | Descriptor: | CALCIUM ION, PHOSPHATE ION, RABPHILIN-3A | | Authors: | Schlicker, C, Montaville, P, Sheldrick, G.M, Becker, S. | | Deposit date: | 2006-05-04 | | Release date: | 2006-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The C2A-C2B Linker Defines the High Affinity Ca2+ Binding Mode of Rabphilin-3A.
J.Biol.Chem., 282, 2007
|
|
7T8G
 
 | |
5EHS
 
 | |
5IQR
 
 | | Structure of RelA bound to the 70S ribosome | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Brown, A, Fernandez, I.S, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-03-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ribosome-dependent activation of stringent control.
Nature, 534, 2016
|
|
5EIA
 
 | | mACHE-anti TZ2PA5 complex from a 1:6 mixture of the syn/anti isomers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-phenyl-5-[5-[1-[2-(1,2,3,4-tetrahydroacridin-9-ylamino)ethyl]-1,2,3-triazol-4-yl]pentyl]phenanthridin-5-ium-3,8-diamine, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Marchot, P. | | Deposit date: | 2015-10-29 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Steric and Dynamic Parameters Influencing In Situ Cycloadditions to Form Triazole Inhibitors with Crystalline Acetylcholinesterase.
J.Am.Chem.Soc., 138, 2016
|
|
7T8E
 
 | | G93A mutant of human SOD1 in P21 space group | | Descriptor: | ACETATE ION, SULFATE ION, Superoxide dismutase [Cu-Zn], ... | | Authors: | Amporndanai, K, Hasnain, S.S. | | Deposit date: | 2021-12-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ebselen analogues delay disease onset and its course in fALS by on-target SOD-1 engagement.
Sci Rep, 14, 2024
|
|
5EJ2
 
 | |
2C6M
 
 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | 4-{[5-(CYCLOHEXYLOXY)[1,2,4]TRIAZOLO[1,5-A]PYRIMIDIN-7-YL]AMINO}BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-10 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Triazolo[1,5-A]Pyrimidines as Novel Cdk2 Inhibitors: Protein Structure-Guided Design and Sar.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4APC
 
 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) | | Descriptor: | CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1 | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
7T8H
 
 | |
5ERV
 
 | |
5LX3
 
 | | CRYSTAL STRUCTURE OF VISFATIN IN COMPLEX WITH SAR154782. | | Descriptor: | 6-[4-[(6-azanylpyridin-3-yl)methylcarbamoylamino]-3-fluoranyl-phenyl]-2-(ethylamino)-~{N}-(2-piperidin-1-ylethyl)pyridine-3-carboxamide, Nicotinamide phosphoribosyltransferase | | Authors: | Bertrand, T, Marquette, J.P. | | Deposit date: | 2016-09-20 | | Release date: | 2017-10-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF VISFATIN IN COMPLEX WITH SAR154782.
To Be Published
|
|
5ELX
 
 | | S. cerevisiae Dbp5 bound to RNA and mant-ADP BeF3 | | Descriptor: | ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Merchant, M.K, Modis, Y. | | Deposit date: | 2015-11-05 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Pi Release Limits the Intrinsic and RNA-Stimulated ATPase Cycles of DEAD-Box Protein 5 (Dbp5).
J.Mol.Biol., 428, 2016
|
|
1SVE
 
 | | Crystal Structure of Protein Kinase A in Complex with Azepane Derivative 1 | | Descriptor: | (4R)-4-(2-FLUORO-6-HYDROXY-3-METHOXY-BENZOYL)-BENZOIC ACID (3R)-3-[(PYRIDINE-4-CARBONYL)AMINO]-AZEPAN-4-YL ESTER, N-OCTANOYL-N-METHYLGLUCAMINE, SODIUM ION, ... | | Authors: | Breitenlechner, C.B, Wegge, T, Berillon, L, Graul, K, Marzenell, K, Friebe, W.-G, Thomas, U, Schumacher, R, Huber, R, Engh, R.A, Masjost, B. | | Deposit date: | 2004-03-29 | | Release date: | 2005-03-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-based optimization of novel azepane derivatives as PKB inhibitors
J.Med.Chem., 47, 2004
|
|
6DWZ
 
 | | Hermes transposase deletion dimer complex with (C/G) DNA | | Descriptor: | DNA (26-MER), DNA (5'-D(*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*C)-3'), ... | | Authors: | Dyda, F, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
