6UDM
 
 | |
8TJH
 
 | |
6ODA
 
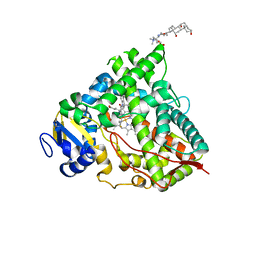
 | | Crystal structure of HDAC8 in complex with compound 2 | | Descriptor: | Histone deacetylase 8, N-{2-[3-(hydroxyamino)-3-oxopropyl]phenyl}-3-(trifluoromethyl)benzamide, POTASSIUM ION, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
6QDX
 
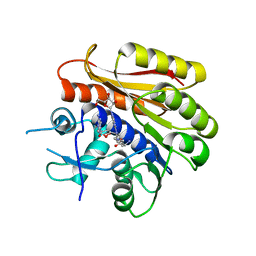 | | Structure of E.coli RlmJ in complex with a bisubstrate analogue (BA4) | | Descriptor: | (2~{S})-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-[3-[[9-[(2~{S},3~{R},4~{S},5~{S})-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]purin-6-yl]amino]propyl]amino]-2-azanyl-butanoic acid, Ribosomal RNA large subunit methyltransferase J | | Authors: | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | Deposit date: | 2019-01-03 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
8FIY
 
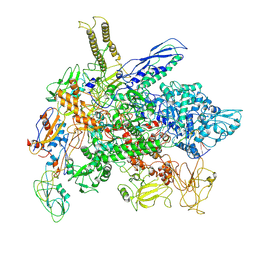 | | Cryo-EM structure of E. coli RNA polymerase Elongation complex in the Transcription-Translation Complex (RNAP in an anti-swiveled conformation) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-17 | | Release date: | 2023-03-29 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
6ODB
 
 | | Crystal structure of HDAC8 in complex with compound 3 | | Descriptor: | GLYCEROL, Histone deacetylase 8, N-{2-[(1E)-3-(hydroxyamino)-3-oxoprop-1-en-1-yl]phenyl}-2-phenoxybenzamide, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
8FIX
 
 | | Cryo-EM structure of E. coli RNA polymerase backtracked elongation complex harboring a terminal mismatch | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Florez Ariza, A, Wee, L, Tong, A, Canari, C, Grob, P, Nogales, E, Bustamante, C. | | Deposit date: | 2022-12-17 | | Release date: | 2023-03-29 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A trailing ribosome speeds up RNA polymerase at the expense of transcript fidelity via force and allostery.
Cell, 186, 2023
|
|
6O97
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with propylamycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.75A resolution | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxyc yclohexyl 2-amino-2,4-dideoxy-4-propyl-alpha-D-glucopyranoside, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Matsushita, T, Sati, G.C, Kondasinghe, N, Pirrone, M.G, Kato, T, Waduge, P, Kumar, H.S, Sanchon, A.C, Dobosz-Bartoszek, M, Shcherbakov, D, Juhas, M, Hobbie, S.N, Schrepfer, T, Chow, C.S, Polikanov, Y.S, Schacht, J, Vasella, A, Bottger, E.C, Crich, D. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-17 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Design, Multigram Synthesis, and in Vitro and in Vivo Evaluation of Propylamycin: A Semisynthetic 4,5-Deoxystreptamine Class Aminoglycoside for the Treatment of Drug-Resistant Enterobacteriaceae and Other Gram-Negative Pathogens.
J. Am. Chem. Soc., 141, 2019
|
|
8ZJF
 
 | | Cryo-EM structure of human integrin alpha-E beta-7 | | Descriptor: | CALCIUM ION, Integrin alpha-E, Integrin beta-7, ... | | Authors: | Akasaka, H, Nureki, O, Kise, Y. | | Deposit date: | 2024-05-14 | | Release date: | 2024-06-05 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structure of I domain-containing integrin alpha E beta 7.
Biochem.Biophys.Res.Commun., 721, 2024
|
|
6NU9
 
 | |
8UD6
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.70A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-thiabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Wu, K.J.Y, Tresco, B.I.C, Ramkissoon, A, See, D.N.Y, Liow, P, Dittemore, G.A, Yu, M, Testolin, G, Mitcheltree, M.J, Liu, R.Y, Svetlov, M.S, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-02-21 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance.
Science, 383, 2024
|
|
8UD7
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.70A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-thiabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Wu, K.J.Y, Tresco, B.I.C, Ramkissoon, A, See, D.N.Y, Liow, P, Dittemore, G.A, Yu, M, Testolin, G, Mitcheltree, M.J, Liu, R.Y, Svetlov, M.S, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-02-21 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance.
Science, 383, 2024
|
|
9DFD
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with lasso peptide lariocidin B, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.60A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Aleksandrova, E.V, Travin, D.Y, Jangra, M, Kaur, M, Darwish, L, Koteva, K, Klepacki, D, Wang, W, Tiffany, M, Sokaribo, A, Coombes, B.K, Vazquez-Laslop, N, Wright, G.D, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2024-08-29 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Broad Spectrum Lasso Peptide Antibiotic Targeting the Bacterial Ribosome.
Res Sq, 2024
|
|
8UD8
 
 | | Crystal structure of the A2503-C2,C8-dimethylated Thermus thermophilus 70S ribosome in complex with cresomycin, mRNA, deacylated A-site tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.70A resolution | | Descriptor: | (4S,5aS,8S,8aR)-4-(2-methylpropyl)-N-[(1R,5Z,7R,8R,9R,10R,11S,12R)-10,11,12-trihydroxy-7-methyl-13-oxa-2-thiabicyclo[7.3.1]tridec-5-en-8-yl]octahydro-2H-oxepino[2,3-c]pyrrole-8-carboxamide (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Syroegin, E.A, Wu, K.J.Y, Tresco, B.I.C, Ramkissoon, A, See, D.N.Y, Liow, P, Dittemore, G.A, Yu, M, Testolin, G, Mitcheltree, M.J, Liu, R.Y, Svetlov, M.S, Myers, A.G, Polikanov, Y.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-02-21 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An antibiotic preorganized for ribosomal binding overcomes antimicrobial resistance.
Science, 383, 2024
|
|
5MF4
 
 | | Tubulin-Dictyostatin complex | | Descriptor: | (3~{Z},5~{E},7~{R},8~{S},10~{S},11~{Z},13~{S},14~{R},15~{S},17~{S},20~{R},21~{S},22~{S})-22-[(2~{S},3~{Z})-hexa-3,5-dien-2-yl]-7,13,15,17,21-pentamethyl-8,10,14,20-tetrakis(oxidanyl)-1-oxacyclodocosa-3,5,11-trien-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Trigili, C, Barasoain, I, Sanchez-Murcia, P.A, Bargsten, K, Redondo-Horcajo, M, Nogales, A, Gardner, N.M, Meyer, A, Naylor, G.J, Gomez-Rubio, E, Gago, F, Steinmetz, M.O, Paterson, I, Prota, A.E, Diaz, J.F. | | Deposit date: | 2016-11-17 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Determinants of the Dictyostatin Chemotype for Tubulin Binding Affinity and Antitumor Activity Against Taxane- and Epothilone-Resistant Cancer Cells.
ACS Omega, 1, 2016
|
|
2JX2
 
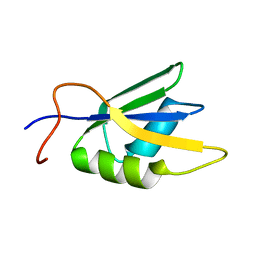 | | Solution conformation of RNA-bound NELF-E RRM | | Descriptor: | Negative elongation factor E | | Authors: | Jampani, N, Schweimer, K, Wenzel, S, Woehrl, B.M, Roesch, P. | | Deposit date: | 2007-11-02 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NELF-E RRM Undergoes Major Structural Changes in Flexible Protein Regions on Target RNA Binding
Biochemistry, 47, 2008
|
|
2KK7
 
 | |
6U7B
 
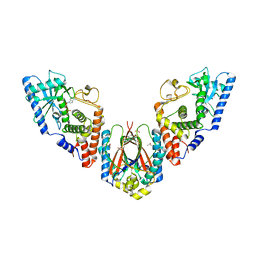 | | Structure of E. coli MS115-1 CdnC:HORMA-deltaN complex | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
7UIB
 
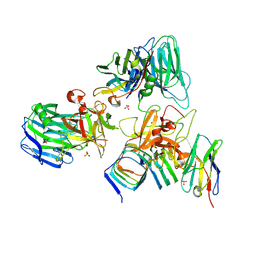 | | Crystal structure of BoNT/E receptor binding domain in complex with SV2, VHH, and sialic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-beta-neuraminic acid, ... | | Authors: | Liu, Z, Jin, R, Chen, P. | | Deposit date: | 2022-03-29 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Nat Commun, 14, 2023
|
|
7UIA
 
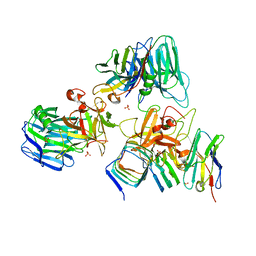 | | Crystal structure of BoNT/E receptor binding domain in complex with SV2 and VHH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Liu, Z, Jin, R, Chen, P. | | Deposit date: | 2022-03-28 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural basis for botulinum neurotoxin E recognition of synaptic vesicle protein 2.
Nat Commun, 14, 2023
|
|
3DHX
 
 | | Crystal structure of isolated C2 domain of the methionine uptake transporter | | Descriptor: | IODIDE ION, Methionine import ATP-binding protein metN | | Authors: | Johnson, E, Kaiser, J.T, Lee, A.T, Rees, D.C. | | Deposit date: | 2008-06-18 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation.
Science, 321, 2008
|
|
3DW7
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2656-SeCH3 modified | | Descriptor: | Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DW6
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-SECH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3DW4
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-OCH3 modified | | Descriptor: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | Authors: | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | Deposit date: | 2008-07-21 | | Release date: | 2009-03-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
6MKF
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
