4QI7
 
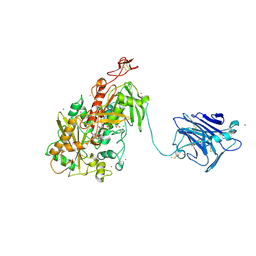 | | Cellobiose dehydrogenase from Neurospora crassa, NcCDH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cellobiose dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
4QI4
 
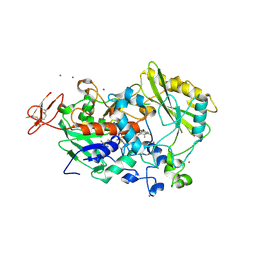 | | Dehydrogenase domain of Myriococcum thermophilum cellobiose dehydrogenase, MtDH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
4QI5
 
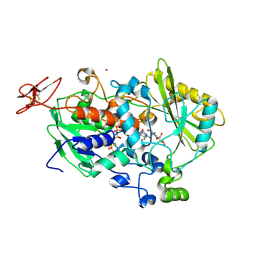 | | Dehydrogenase domain of Myriococcum thermophilum cellobiose dehydrogenase with bound cellobionolactam, MtDH | | Descriptor: | (2R,3R,4R,5R)-4,5-dihydroxy-2-(hydroxymethyl)-6-oxopiperidin-3-yl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
4QI8
 
 | | Lytic polysaccharide monooxygenase 9F from Neurospora crassa, NcLPMO9F | | Descriptor: | COPPER (II) ION, Lytic polysaccharide monooxygenase, NITRATE ION | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
4QI3
 
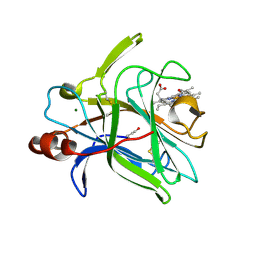 | | Cytochrome domain of Myriococcum thermophilum cellobiose dehydrogenase, MtCYT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cellobiose dehydrogenase, MAGNESIUM ION, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
4QI6
 
 | | Cellobiose dehydrogenase from Myriococcum thermophilum, MtCDH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cellobiose dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Gandini, R, Sygmund, C, Kittl, R, Haltrich, D, Ludwig, R, Hallberg, B.M, Divne, C. | | Deposit date: | 2014-05-30 | | Release date: | 2015-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for cellobiose dehydrogenase action during oxidative cellulose degradation.
Nat Commun, 6, 2015
|
|
3FDY
 
 | | Pyranose 2-oxidase thermostable triple mutant, T169G/E542K/V546C | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose oxidase (Pyranose 2-oxidase) | | Authors: | Tan, T.C, Divne, C. | | Deposit date: | 2008-11-26 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A thermostable triple mutant of pyranose 2-oxidase from Trametes multicolor with improved properties for biotechnological applications
Biotechnol J, 4, 2009
|
|
4H7U
 
 | | Crystal structure of pyranose dehydrogenase from Agaricus meleagris, wildtype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PHOSPHATE ION, ... | | Authors: | Tan, T.C, Spadiut, O, Divne, C. | | Deposit date: | 2012-09-20 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A crystal structure of pyranose dehydrogenase from Agaricus meleagris rationalizes substrate specificity and reveals a flavin intermediate.
Plos One, 8, 2013
|
|
3BG6
 
 | | Pyranose 2-oxidase from Trametes multicolor, E542K mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyranose oxidase | | Authors: | Tan, T.C, Divne, C. | | Deposit date: | 2007-11-26 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Improving thermostability and catalytic activity of pyranose 2-oxidase from Trametes multicolor by rational and semi-rational design
Febs J., 276, 2009
|
|
3LSK
 
 | | Pyranose 2-oxidase T169S acetate complex | | Descriptor: | ACETATE ION, DODECAETHYLENE GLYCOL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Spadiut, O, Divne, C. | | Deposit date: | 2010-02-12 | | Release date: | 2010-08-25 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | H-bonding and positive charge at the N5/O4 locus are critical for covalent flavin attachment in trametes pyranose 2-oxidase.
J.Mol.Biol., 402, 2010
|
|
6LNU
 
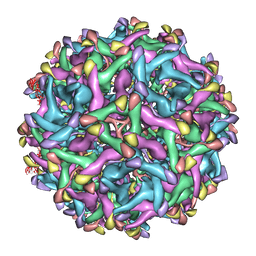 | | Cryo-EM structure of immature Zika virus | | Descriptor: | Genome polyprotein | | Authors: | Tan, T.Y, Fibriansah, G, Kostyuchenko, V.A, Ng, T.S, Lim, X.X, Lim, X.N, Shi, J, Morais, M.C, Corti, D, Lok, S.M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Capsid protein structure in Zika virus reveals the flavivirus assembly process.
Nat Commun, 11, 2020
|
|
6LNT
 
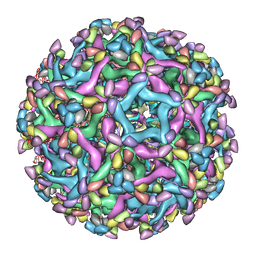 | | Cryo-EM structure of immature Zika virus in complex with human antibody DV62.5 Fab | | Descriptor: | Envelope protein, Fab DV62.5 heavy-chain variable region, Fab DV62.5 light-chain variable region, ... | | Authors: | Tan, T.Y, Fibriansah, G, Kostyuchenko, V.A, Ng, T.S, Lim, X.X, Lim, X.N, Shi, J, Morais, M.C, Corti, D, Lok, S.M. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Capsid protein structure in Zika virus reveals the flavivirus assembly process.
Nat Commun, 11, 2020
|
|
3LSI
 
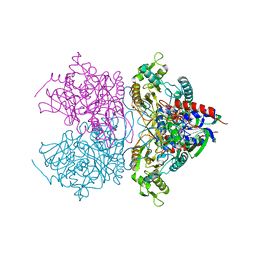 | | Pyranose 2-oxidase T169A, tetragonal | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyranose 2-oxidase | | Authors: | Tan, T.C, Spadiut, O, Divne, C. | | Deposit date: | 2010-02-12 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | H-bonding and positive charge at the N5/O4 locus are critical for covalent flavin attachment in trametes pyranose 2-oxidase.
J.Mol.Biol., 402, 2010
|
|
3LSM
 
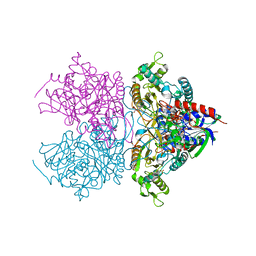 | | Pyranose 2-oxidase H167A mutant with flavin N(5) sulfite adduct | | Descriptor: | (S)-10-((2S,3S,4R)-5-((S)-((S)-(((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHOXY)(HYDROXY)PHOSPHORYLOXY)(HYDROXY)PHOSPHORYLOXY)-2,3,4-TRIHYDROXYPENTYL)-7,8-DIMETHYL-2,4-DIOXO-2,3,4,4A-TETRAHYDROBENZO[G]PTERIDINE-5(10H)-SULFONIC ACID, DODECAETHYLENE GLYCOL, Pyranose 2-oxidase, ... | | Authors: | Tan, T.C, Spadiut, O, Divne, C. | | Deposit date: | 2010-02-12 | | Release date: | 2010-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | H-bonding and positive charge at the N5/O4 locus are critical for covalent flavin attachment in trametes pyranose 2-oxidase.
J.Mol.Biol., 402, 2010
|
|
4MOL
 
 | | Pyranose 2-oxidase H167A mutant with 2-fluorinated galactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-beta-D-galactopyranose, DODECAETHYLENE GLYCOL, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
4MOJ
 
 | | Pyranose 2-oxidase H450G/V546C double mutant with 2-fluorinated glucose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-alpha-D-glucopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
4MOP
 
 | | Pyranose 2-oxidase V546C mutant with 3-fluorinated galactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-deoxy-3-fluoro-beta-D-galactopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
4MOM
 
 | | Pyranose 2-oxidase H450G mutant with 3-fluorinated galactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-deoxy-3-fluoro-beta-D-galactopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
4MOE
 
 | | Pyranose 2-oxidase H450G mutant with 3-fluorinated glucose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-deoxy-3-fluoro-beta-D-glucopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
4MOR
 
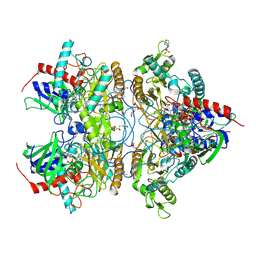 | | Pyranose 2-oxidase H450G/V546C double mutant with 3-fluorinated galactose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-deoxy-3-fluoro-beta-D-galactopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
3BC9
 
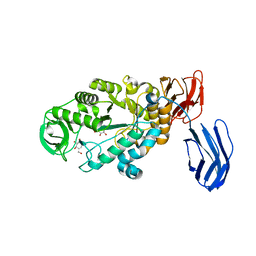 | | Alpha-amylase B in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
3BCD
 
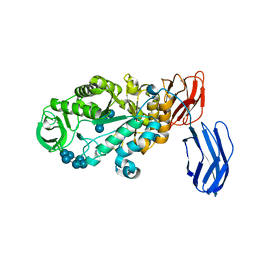 | | Alpha-amylase B in complex with maltotetraose and alpha-cyclodextrin | | Descriptor: | Alpha amylase, catalytic region, CALCIUM ION, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
3BCF
 
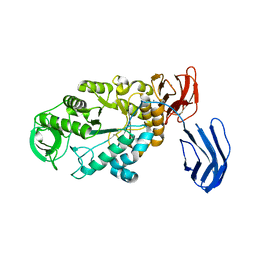 | | Alpha-amylase B from Halothermothrix orenii | | Descriptor: | Alpha amylase, catalytic region, CALCIUM ION, ... | | Authors: | Tan, T.-C, Mijts, B.N, Swaminathan, K, Patel, B.K.C, Divne, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Polyextremophilic alpha-Amylase AmyB from Halothermothrix orenii: Details of a Productive Enzyme-Substrate Complex and an N Domain with a Role in Binding Raw Starch
J.Mol.Biol., 378, 2008
|
|
3BLY
 
 | | Pyranose 2-oxidase from Trametes multicolor, E542K/L537W | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose oxidase | | Authors: | Tan, T.C, Divne, C. | | Deposit date: | 2007-12-11 | | Release date: | 2008-12-02 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Improving thermostability and catalytic activity of pyranose 2-oxidase from Trametes multicolor by rational and semi-rational design
Febs J., 276, 2009
|
|
4MOH
 
 | | Pyranose 2-oxidase V546C mutant with 2-fluorinated glucose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-beta-D-glucopyranose, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, ... | | Authors: | Tan, T.C, Spadiut, O, Gandini, R, Haltrich, D, Divne, C. | | Deposit date: | 2013-09-12 | | Release date: | 2014-02-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Binding of Fluorinated Glucose and Galactose to Trametes multicolor Pyranose 2-Oxidase Variants with Improved Galactose Conversion.
Plos One, 9, 2014
|
|
