3U5O
 
 | |
3U5P
 
 | |
3U5N
 
 | |
6H8P
 
 | | JMJD2A/ KDM4A COMPLEXED WITH NI(II), NOG AND Histone H1.4(18-32)K26me3 peptide (15-mer) | | Descriptor: | CHLORIDE ION, GLYCEROL, Histone H1.4, ... | | Authors: | Chowdhury, R, Walport, L.J, Schofield, C.J. | | Deposit date: | 2018-08-03 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | Mechanistic and structural studies of KDM-catalysed demethylation of histone 1 isotype 4 at lysine 26.
FEBS Lett., 592, 2018
|
|
6B9M
 
 | |
3T6R
 
 | | Structure of UHRF1 in complex with unmodified H3 N-terminal tail | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3.1t N-terminal peptide, MAGNESIUM ION, ... | | Authors: | Xie, S, Jakoncic, J, Qian, C.M. | | Deposit date: | 2011-07-29 | | Release date: | 2011-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | UHRF1 double tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail
J.Mol.Biol., 415, 2012
|
|
3DB4
 
 | | Crystal structure of the tandem tudor domains of the E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, SULFATE ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Dong, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein.
J.Biol.Chem., 286, 2011
|
|
3LWE
 
 | | The crystal structure of MPP8 | | Descriptor: | M-phase phosphoprotein 8 | | Authors: | Li, Z, Li, Z, Ruan, J, Xu, C, Tong, Y, Pan, P.W, Tempel, W, Crombet, L, Min, J, Zang, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-23 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for specific binding of human MPP8 chromodomain to histone H3 methylated at lysine 9.
Plos One, 6, 2011
|
|
3DB3
 
 | | Crystal structure of the tandem tudor domains of the E3 ubiquitin-protein ligase UHRF1 in complex with trimethylated histone H3-K9 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Trimethylated histone H3-K9 peptide | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Dong, A, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-05-30 | | Release date: | 2008-09-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein.
J.Biol.Chem., 286, 2011
|
|
4QUC
 
 | | Crystal structure of chromodomain of Rhino | | Descriptor: | RE36324p | | Authors: | Li, S, Patel, D.J. | | Deposit date: | 2014-07-10 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing.
Genes Dev., 28, 2014
|
|
3SVM
 
 | | Human MPP8 - human DNMT3AK47me2 peptide | | Descriptor: | DNA (cytosine-5)-methyltransferase 3A, M-phase phosphoprotein 8 | | Authors: | Chang, Y, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2011-07-12 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a.
Nat Commun, 2, 2011
|
|
2L3R
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Histone H3 peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Histone H3 | | Authors: | Nady, N, Lemak, A, Fares, C, Gutmanas, A, Avvakumov, G, Xue, S, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-21 | | Release date: | 2011-04-13 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Recognition of Multivalent Histone States Associated with Heterochromatin by UHRF1 Protein.
J.Biol.Chem., 286, 2011
|
|
3K3O
 
 | | Crystal structure of the catalytic core domain of human PHF8 complexed with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, PHD finger protein 8 | | Authors: | Yu, L, Wang, Y, Huang, S, Wang, J, Deng, Z, Wu, W, Gong, W, Chen, Z. | | Deposit date: | 2009-10-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into a novel histone demethylase PHF8
Cell Res., 20, 2010
|
|
5FB1
 
 | | Crystal Structure of a PHD finger bound to histone H3 K9me3 peptide | | Descriptor: | MALONATE ION, Nuclear autoantigen Sp-100, Peptide from Histone H3, ... | | Authors: | Li, H, Zhang, X. | | Deposit date: | 2015-12-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multifaceted Histone H3 Methylation and Phosphorylation Readout by the Plant Homeodomain Finger of Human Nuclear Antigen Sp100C
J.Biol.Chem., 291, 2016
|
|
5FB0
 
 | | Crystal Structure of a PHD finger bound to histone H3 T3ph peptide | | Descriptor: | Nuclear autoantigen Sp-100, Peptide from Histone H3, ZINC ION | | Authors: | Li, H, Zhang, X. | | Deposit date: | 2015-12-13 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Multifaceted Histone H3 Methylation and Phosphorylation Readout by the Plant Homeodomain Finger of Human Nuclear Antigen Sp100C
J.Biol.Chem., 291, 2016
|
|
3K3N
 
 | | Crystal structure of the catalytic core domain of human PHF8 | | Descriptor: | FE (II) ION, PHD finger protein 8 | | Authors: | Yu, L, Wang, Y, Huang, S, Wang, J, Deng, Z, Wu, W, Gong, W, Chen, Z. | | Deposit date: | 2009-10-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into a novel histone demethylase PHF8
Cell Res., 20, 2010
|
|
6F6D
 
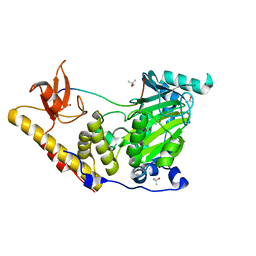 | | The catalytic domain of KDM6B in complex with H3(17-33)K18IA21M peptide | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Histone 3 peptide H3(17-33)K18IA21M, ... | | Authors: | Jones, S.E, Olsen, L, Gajhede, M. | | Deposit date: | 2017-12-05 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81810677 Å) | | Cite: | Structural Basis of Histone Demethylase KDM6B Histone 3 Lysine 27 Specificity.
Biochemistry, 57, 2018
|
|
5OY3
 
 | | The structural basis of the histone demethylase KDM6B histone 3 lysine 27 specificity | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FE (III) ION, ... | | Authors: | Jones, S.E, Olsen, L, Gajhede, M. | | Deposit date: | 2017-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.136 Å) | | Cite: | Structural Basis of Histone Demethylase KDM6B Histone 3 Lysine 27 Specificity.
Biochemistry, 57, 2018
|
|
7Z36
 
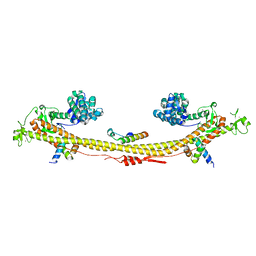 | | Crystal structure of the KAP1 tripartite motif in complex with the ZNF93 KRAB domain | | Descriptor: | Endolysin,Transcription intermediary factor 1-beta,Isoform 2 of Transcription intermediary factor 1-beta, SMARCAD1 CUE1 domain, ZINC ION, ... | | Authors: | Stoll, G.A, Modis, Y. | | Deposit date: | 2022-03-01 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and functional mapping of the KRAB-KAP1 repressor complex.
Embo J., 41, 2022
|
|
4V2V
 
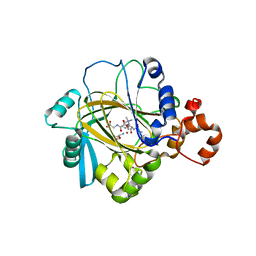 | | JMJD2A COMPLEXED WITH NI(II), NOG AND HISTONE H3K27me3 PEPTIDE (25-29) ARK(me3)SA | | Descriptor: | CHLORIDE ION, HISTONE H3.1T, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Chowdhury, R, Madden, S.K, Schofield, C.J. | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Studies on the Catalytic Domains of Multiple Jmjc Oxygenases Using Peptide Substrates.
Epigenetics, 9, 2014
|
|
4V2W
 
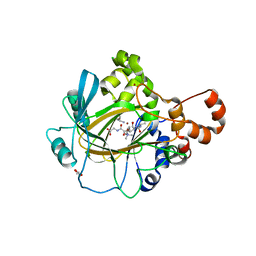 | | JMJD2A COMPLEXED WITH NI(II), NOG AND HISTONE H3K27me3 PEPTIDE (16-35) | | Descriptor: | CHLORIDE ION, GLYCEROL, HISTONE H3.1T, ... | | Authors: | Chowdhury, R, Zafred, D, Schofield, C.J. | | Deposit date: | 2014-10-15 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Studies on the Catalytic Domains of Multiple Jmjc Oxygenases Using Peptide Substrates.
Epigenetics, 9, 2014
|
|
6QAJ
 
 | | Structure of the tripartite motif of KAP1/TRIM28 | | Descriptor: | Endolysin,Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Stoll, G.A, Oda, S, Yu, M, Modis, Y. | | Deposit date: | 2018-12-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structure of KAP1 tripartite motif identifies molecular interfaces required for retroelement silencing.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6BHH
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, SULFATE ION, ... | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
5VGI
 
 | | Crystal Structure of KDM4 with the Small Molecule Inhibitor QC6352 | | Descriptor: | 3-[({(1R)-6-[methyl(phenyl)amino]-1,2,3,4-tetrahydronaphthalen-1-yl}methyl)amino]pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Hosfield, D.J. | | Deposit date: | 2017-04-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Design of KDM4 Inhibitors with Antiproliferative Effects in Cancer Models.
ACS Med Chem Lett, 8, 2017
|
|
5VMP
 
 | | Crystal Structure of Human KDM4 with Small Molecule Inhibitor QC5714 | | Descriptor: | 3-({[(1R)-6-methoxy-1,2,3,4-tetrahydronaphthalen-1-yl]methyl}amino)pyridine-4-carboxylic acid, Lysine-specific demethylase 4A, NICKEL (II) ION, ... | | Authors: | Hosfield, D.J. | | Deposit date: | 2017-04-28 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Design of KDM4 Inhibitors with Antiproliferative Effects in Cancer Models.
ACS Med Chem Lett, 8, 2017
|
|
