1YW2
 
 | | Mutated Mus Musculus P38 Kinase (mP38) | | Descriptor: | 2-(ETHOXYMETHYL)-4-(4-FLUOROPHENYL)-3-[2-(2-HYDROXYPHENOXY)PYRIMIDIN-4-YL]ISOXAZOL-5(2H)-ONE, Mitogen-activated protein kinase 14 | | Authors: | Laughlin, S.K, Clark, M.P, Djung, J.F, Golebiowski, A, Brugel, T.A, Sabat, M, Bookland, R.G, Laufersweiler, M.J, Vanrens, J.C, Townes, J.A, De, B, Hsieh, L.C, Xu, S.C, Walter, R.L, Mekel, M.J, Janusz, M.J. | | Deposit date: | 2005-02-16 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The development of new isoxazolone based inhibitors of tumor necrosis factor-alpha (TNF-alpha) production.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2Z7L
 
 | |
1YWR
 
 | | Crystal Structure Analysis of inactive P38 kinase domain in complex with a Monocyclic Pyrazolone Inhibitor | | Descriptor: | 4-(4-FLUOROPHENYL)-1-METHYL-5-(2-{[(1S)-1-PHENYLETHYL]AMINO}PYRIMIDIN-4-YL)-2-PIPERIDIN-4-YL-1,2-DIHYDRO-3H-PYRAZOL-3-ONE, Mitogen-activated protein kinase 14 | | Authors: | Golebiowski, A, Townes, J.A, Laufersweiler, M.J, Brugel, T.A, Clark, M.P, Clark, C.M, Djung, J.F, Laughlin, S.K, Sabat, M.P, Bookland, R.G, Vanrens, J.C, De, B, Hsieh, L.C, Janusz, M.J, Walter, R.L, Webster, M.E, Mekel, M.J. | | Deposit date: | 2005-02-18 | | Release date: | 2005-05-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The development of monocyclic pyrazolone based cytokine synthesis inhibitors.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
3B3A
 
 | | Structure of E163K/R145E DJ-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Protein DJ-1 | | Authors: | Lakshminarasimhan, M, Maldonado, M.T, Zhou, W, Fink, A.L, Wilson, M.A. | | Deposit date: | 2007-10-19 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Impact of Three Parkinsonism-Associated Missense Mutations on Human DJ-1.
Biochemistry, 47, 2008
|
|
3C9W
 
 | | Crystal Structure of ERK-2 with hypothemycin covalently bound | | Descriptor: | (1aR,8S,13S,14S,15aR)-5,13,14-trihydroxy-3-methoxy-8-methyl-8,9,13,14,15,15a-hexahydro-6H-oxireno[k][2]benzoxacyclotetradecine-6,12(1aH)-dione, Mitogen-activated protein kinase 1 | | Authors: | Rosenfeld, R.J. | | Deposit date: | 2008-02-18 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular modeling and crystal structure of ERK2-hypothemycin complexes
J.Struct.Biol., 164, 2008
|
|
4I6M
 
 | | Structure of Arp7-Arp9-Snf2(HSA)-RTT102 subcomplex of SWI/SNF chromatin remodeler. | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, PHOSPHATE ION, ... | | Authors: | Schubert, H.L, Cairns, B.R, Hill, C.P. | | Deposit date: | 2012-11-29 | | Release date: | 2013-02-13 | | Last modified: | 2017-08-16 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structure of an actin-related subcomplex of the SWI/SNF chromatin remodeler.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7Z6O
 
 | | X-Ray studies of Ku70/80 reveal the binding site for IP6 | | Descriptor: | DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*AP*AP*AP*CP*TP*AP*AP*AP*AP*AP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Varela, P.F, Charbonnier, J.B. | | Deposit date: | 2022-03-14 | | Release date: | 2023-08-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
1N39
 
 | | Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase | | Descriptor: | CALCIUM ION, DNA complement strand, DNA inhibitor strand, ... | | Authors: | Norman, D.P, Chung, S.J, Verdine, G.L. | | Deposit date: | 2002-10-25 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase
Biochemistry, 42, 2003
|
|
1N3A
 
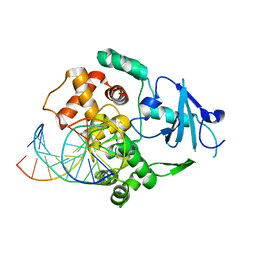 | | Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase | | Descriptor: | CALCIUM ION, DNA complement strand, DNA inhibitor strand, ... | | Authors: | Norman, D.P, Chung, S.J, Verdine, G.L. | | Deposit date: | 2002-10-25 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase
Biochemistry, 42, 2003
|
|
3SMR
 
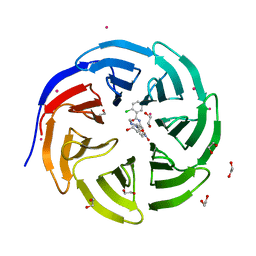 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dombrovski, L, Wasney, G.A, Tempel, W, Senisterra, G, Bolshan, Y, Smil, D, Nguyen, K.T, Hajian, T, Poda, G, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
7ZT6
 
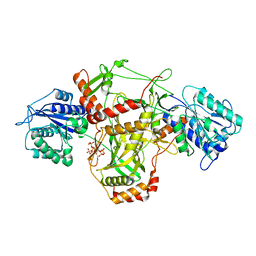 | |
7ZVT
 
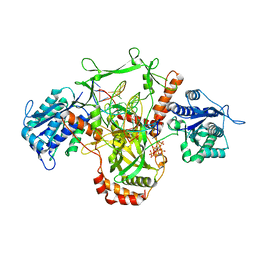 | | CryoEM structure of Ku heterodimer bound to DNA | | Descriptor: | DNA (5'-D(P*CP*GP*AP*TP*AP*TP*CP*TP*AP*GP*AP*GP*GP*GP*AP*T)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*CP*TP*AP*GP*AP*TP*AP*TP*C)-3'), INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-17 | | Release date: | 2023-05-24 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural and functional basis of inositol hexaphosphate stimulation of NHEJ through stabilization of Ku-XLF interaction.
Nucleic Acids Res., 51, 2023
|
|
8CTB
 
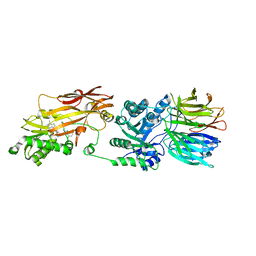 | | Human PRMT5:MEP50 structure with Fragment 3 and MTA Bound | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 7-chloro-1-methyl-1H-benzimidazol-2-amine, Methylosome protein 50, ... | | Authors: | Gunn, R.J, Lawson, J.D, Smith, C.R. | | Deposit date: | 2022-05-13 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Fragment optimization and elaboration strategies - the discovery of two lead series of PRMT5/MTA inhibitors from five fragment hits.
Rsc Med Chem, 13, 2022
|
|
1DY8
 
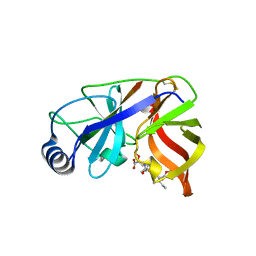 | | Inhibition of the Hepatitis C Virus NS3/4A Protease. The Crystal Structures of Two Protease-Inhibitor Complexes (inhibitor II) | | Descriptor: | N-[(benzyloxy)carbonyl]-L-isoleucyl-N-[(1R)-1-(carboxycarbonyl)-3,3-difluoropropyl]-L-leucinamide, NONSTRUCTURAL PROTEIN NS4A (P4), PROTEASE/HELICASE NS3 (P70) | | Authors: | Di Marco, S, Rizzi, M, Volpari, C, Walsh, M, Narjes, F, Colarusso, S, De Francesco, R, Matassa, V.G, Sollazzo, M. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-28 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Hepatitis C Virus Ns3/4A Protease the Crystal Structures of Two Protease-Inhibitor Complexes
J.Biol.Chem., 275, 2000
|
|
6U62
 
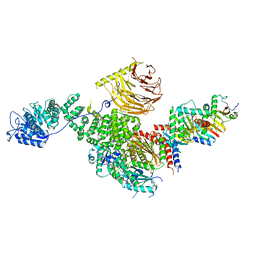 | | Raptor-Rag-Ragulator complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rogala, K.B, Sabatini, D.M. | | Deposit date: | 2019-08-29 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis for the docking of mTORC1 on the lysosomal surface.
Science, 366, 2019
|
|
6QC0
 
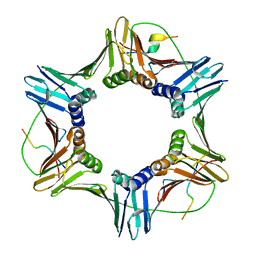 | | PCNA complex with Cdt2 C-terminal PIP-box peptide | | Descriptor: | Denticleless protein homolog, Proliferating cell nuclear antigen | | Authors: | Perrakis, A.P, von Castelmur, E. | | Deposit date: | 2018-12-25 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Direct binding of Cdt2 to PCNA is important for targeting the CRL4Cdt2E3 ligase activity to Cdt1.
Life Sci Alliance, 1, 2018
|
|
6QCG
 
 | |
3S7D
 
 | |
8CYI
 
 | | Cryo-EM structures and computational analysis for enhanced potency in MTA-synergic inhibition of human protein arginine methyltransferase 5 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, N-[(2-aminoquinolin-7-yl)methyl]-9-(2-hydroxyethyl)-2,3,4,9-tetrahydro-1H-carbazole-6-carboxamide, ... | | Authors: | Yadav, G.P, Wei, Z, Xiaozhi, Y, Chenglong, L, Jiang, Q. | | Deposit date: | 2022-05-23 | | Release date: | 2023-04-12 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structure-based selection of computed ligand poses enables design of MTA-synergic PRMT5 inhibitors of better potency.
Commun Biol, 5, 2022
|
|
1LEZ
 
 | | CRYSTAL STRUCTURE OF MAP KINASE P38 COMPLEXED TO THE DOCKING SITE ON ITS ACTIVATOR MKK3B | | Descriptor: | MAP kinase kinase 3b, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Chang, C.-I, Xu, B.-E, Akella, R, Cobb, M.H, Goldsmith, E.J. | | Deposit date: | 2002-04-10 | | Release date: | 2002-07-10 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of MAP kinase p38 complexed to the docking sites on its nuclear substrate MEF2A and activator MKK3b.
Mol.Cell, 9, 2002
|
|
8X3R
 
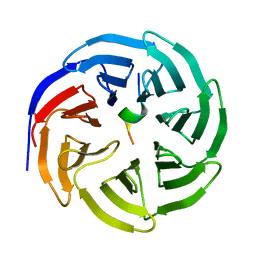 | | Crystal structure of human WDR5 in complex with WDR5 | | Descriptor: | WD repeat-containing protein 5 | | Authors: | Liu, Y, Huang, X. | | Deposit date: | 2023-11-14 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The NTE domain of PTEN alpha / beta promotes cancer progression by interacting with WDR5 via its SSSRRSS motif.
Cell Death Dis, 15, 2024
|
|
8X77
 
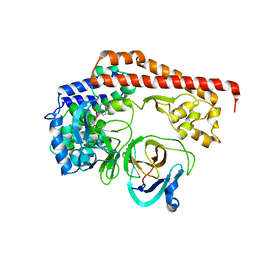 | | Enterovirus proteinase with host factor | | Descriptor: | 2A protein, Actin-histidine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2023-11-23 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | The EV71 2A protease occupies the central cleft of SETD3 and disrupts SETD3-actin interaction.
Nat Commun, 15, 2024
|
|
8X3S
 
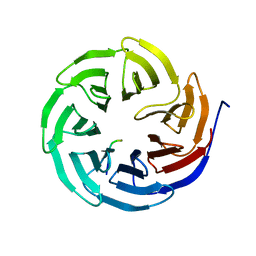 | | Crystal structure of human WDR5 in complex with PTEN | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN, WD repeat-containing protein 5 | | Authors: | Liu, Y, Huang, X, Shang, X. | | Deposit date: | 2023-11-14 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The NTE domain of PTEN alpha / beta promotes cancer progression by interacting with WDR5 via its SSSRRSS motif.
Cell Death Dis, 15, 2024
|
|
8X8Q
 
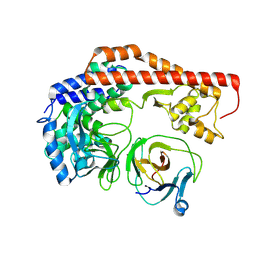 | | Structure of enterovirus protease in complex host factor | | Descriptor: | 2A protein (Fragment), Actin-histidine N-methyltransferase, ZINC ION | | Authors: | Gao, X, Cui, S. | | Deposit date: | 2023-11-28 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The EV71 2A protease occupies the central cleft of SETD3 and disrupts SETD3-actin interaction.
Nat Commun, 15, 2024
|
|
3S7B
 
 | | Structural Basis of Substrate Methylation and Inhibition of SMYD2 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, N-cyclohexyl-N~3~-[2-(3,4-dichlorophenyl)ethyl]-N-(2-{[2-(5-hydroxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-8-yl)ethyl]amino}ethyl)-beta-alaninamide, N-lysine methyltransferase SMYD2, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2011-05-26 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of Substrate Methylation and Inhibition of SMYD2.
Structure, 19, 2011
|
|
