3HKZ
 
 | | The X-ray crystal structure of RNA polymerase from Archaea | | Descriptor: | DNA-directed RNA polymerase subunit 13, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Murakami, K.S. | | Deposit date: | 2009-05-26 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The X-ray crystal structure of RNA polymerase from Archaea.
Nature, 451, 2008
|
|
3LKD
 
 | | Crystal Structure of the type I restriction-modification system methyltransferase subunit from Streptococcus thermophilus, Northeast Structural Genomics Consortium Target SuR80 | | Descriptor: | Type I restriction-modification system methyltransferase subunit | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Mao, M, Xiao, R, Foote, E.L, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-27 | | Release date: | 2010-02-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Northeast Structural Genomics Consortium Target SuR80
To be published
|
|
3KHK
 
 | |
6ESQ
 
 | | Structure of the acetoacetyl-CoA thiolase/HMG-CoA synthase complex from Methanothermococcus thermolithotrophicus soaked with acetyl-CoA | | Descriptor: | CHLORIDE ION, COENZYME A, HydroxyMethylGlutaryl-CoA synthase, ... | | Authors: | Voegeli, B, Engilberge, S, Girard, E, Riobe, F, Maury, O, Erb, J.T, Shima, S, Wagner, T. | | Deposit date: | 2017-10-24 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Archaeal acetoacetyl-CoA thiolase/HMG-CoA synthase complex channels the intermediate via a fused CoA-binding site.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6EXN
 
 | | Post-catalytic P complex spliceosome with 3' splice site docked | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Intron lariat: UBC4 RNA, ... | | Authors: | Wilkinson, M.E, Fica, S.M, Galej, W.P, Norman, C.M, Newman, A.J, Nagai, K. | | Deposit date: | 2017-11-08 | | Release date: | 2018-01-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Postcatalytic spliceosome structure reveals mechanism of 3'-splice site selection.
Science, 358, 2017
|
|
6FBZ
 
 | | Crystal structure of the eIF4E-eIF4G complex from Chaetomium thermophilum in the cap-bound state | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E-like protein,Eukaryotic translation initiation factor 4E-like protein, Eukaryotic translation initiation factor 4G | | Authors: | Gruener, S, Valkov, E. | | Deposit date: | 2017-12-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Structural motifs in eIF4G and 4E-BPs modulate their binding to eIF4E to regulate translation initiation in yeast.
Nucleic Acids Res., 46, 2018
|
|
6ET9
 
 | | Structure of the acetoacetyl-CoA-thiolase/HMG-CoA-synthase complex from Methanothermococcus thermolithotrophicus at 2.75 A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acetyl-CoA acetyltransferase thiolase, CHLORIDE ION, ... | | Authors: | Engilberge, S, Voegeli, B, Girard, E, Riobe, F, Maury, O, Erb, T.J, Shima, S, Wagner, T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Archaeal acetoacetyl-CoA thiolase/HMG-CoA synthase complex channels the intermediate via a fused CoA-binding site.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6FT6
 
 | | Structure of the Nop53 pre-60S particle bound to the exosome nuclear cofactors | | Descriptor: | 25S ribosomal RNA, 5S ribosomal RNA, 60S ribosomal protein L11-A, ... | | Authors: | Schuller, J.M, Falk, S, Conti, E. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-28 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the nuclear exosome captured on a maturing preribosome.
Science, 360, 2018
|
|
6FF4
 
 | | human Bact spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Haselbach, D, Komarov, I, Agafonov, D, Hartmuth, K, Graf, B, Kastner, B, Luehrmann, R, Stark, H. | | Deposit date: | 2018-01-03 | | Release date: | 2018-08-29 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and Conformational Dynamics of the Human Spliceosomal BactComplex.
Cell, 172, 2018
|
|
6FC0
 
 | | Crystal structure of the eIF4E-eIF4G complex from Chaetomium thermophilum | | Descriptor: | Eukaryotic translation initiation factor 4E-like protein, Eukaryotic translation initiation factor 4G | | Authors: | Gruener, S, Valkov, E. | | Deposit date: | 2017-12-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.293 Å) | | Cite: | Structural motifs in eIF4G and 4E-BPs modulate their binding to eIF4E to regulate translation initiation in yeast.
Nucleic Acids Res., 46, 2018
|
|
6G90
 
 | | Prespliceosome structure provides insight into spliceosome assembly and regulation (map A2) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, Cold sensitive U2 snRNA suppressor 1, Pre-mRNA-processing factor 39, ... | | Authors: | Plaschka, C, Lin, P.-C, Charenton, C, Nagai, K. | | Deposit date: | 2018-04-10 | | Release date: | 2018-08-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Prespliceosome structure provides insights into spliceosome assembly and regulation.
Nature, 559, 2018
|
|
7VPG
 
 | | Crystal structure of the C-terminal tail of SARS-CoV-1 Orf6 complex with human nucleoporin pair Rae1-Nup98 | | Descriptor: | Isoform 3 of Nuclear pore complex protein Nup98-Nup96, ORF6 protein, mRNA export factor | | Authors: | Li, T, Guo, H, Yang, T, Wen, Y, Ji, X. | | Deposit date: | 2021-10-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular Mechanism of SARS-CoVs Orf6 Targeting the Rae1-Nup98 Complex to Compete With mRNA Nuclear Export.
Front Mol Biosci, 8, 2021
|
|
7VPH
 
 | | Crystal structure of the C-terminal tail of SARS-CoV-2 Orf6 complex with human nucleoporin pair Rae1-Nup98 | | Descriptor: | Isoform 3 of Nuclear pore complex protein Nup98-Nup96, ORF6 protein, mRNA export factor | | Authors: | Li, T, Guo, H, Yang, T, Wen, Y, Ji, X. | | Deposit date: | 2021-10-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Mechanism of SARS-CoVs Orf6 Targeting the Rae1-Nup98 Complex to Compete With mRNA Nuclear Export.
Front Mol Biosci, 8, 2021
|
|
7VCI
 
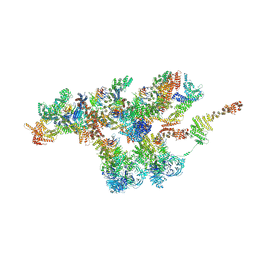 | | Structure of Xenopus laevis NPC nuclear ring asymmetric unit | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Tai, L, Zhu, Y, Sun, F. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
5LJ3
 
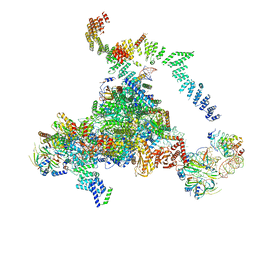 | | Structure of the core of the yeast spliceosome immediately after branching | | Descriptor: | CEF1, CLF1, CWC15, ... | | Authors: | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-03 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
7WB4
 
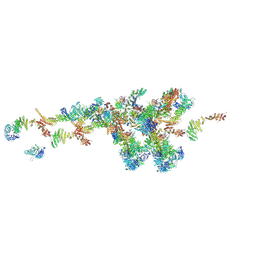 | | Cryo-EM structure of the NR subunit from X. laevis NPC | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Huang, G, Zhan, X, Shi, Y. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structure of the nuclear ring from Xenopus laevis nuclear pore complex.
Cell Res., 32, 2022
|
|
7W5B
 
 | | The cryo-EM structure of human C* complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanism of exon ligation by human spliceosome.
Mol.Cell, 82, 2022
|
|
7W5A
 
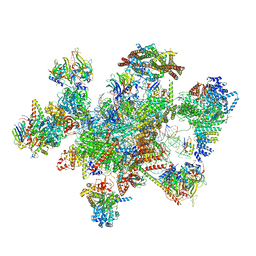 | | The cryo-EM structure of human pre-C*-II complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of exon ligation by human spliceosome.
Mol.Cell, 82, 2022
|
|
7W59
 
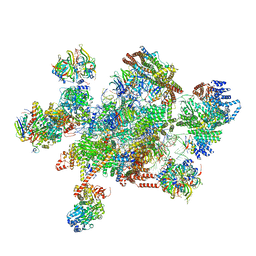 | | The cryo-EM structure of human pre-C*-I complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2021-11-29 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanism of exon ligation by human spliceosome.
Mol.Cell, 82, 2022
|
|
5MPS
 
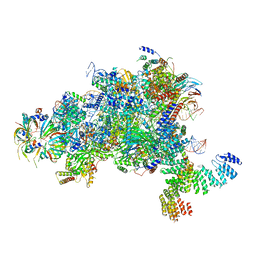 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-18 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
5MQ0
 
 | | Structure of a spliceosome remodeled for exon ligation | | Descriptor: | 3'-EXON OF UBC4 PRE-MRNA, BOUND BY PRP22 HELICASE, 5'-EXON OF UBC4 PRE-MRNA, ... | | Authors: | Fica, S.M, Oubridge, C, Galej, W.P, Wilkinson, M.E, Newman, A.J, Bai, X.-C, Nagai, K. | | Deposit date: | 2016-12-19 | | Release date: | 2017-01-18 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (4.17 Å) | | Cite: | Structure of a spliceosome remodelled for exon ligation.
Nature, 542, 2017
|
|
5LJ5
 
 | | Overall structure of the yeast spliceosome immediately after branching. | | Descriptor: | CWC15, CWC22, Exon 1 (5' exon) of UBC4 pre-mRNA, ... | | Authors: | Galej, W.P, Wilkinson, M.F, Fica, S.M, Oubridge, C, Newman, A.J, Nagai, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-31 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the spliceosome immediately after branching.
Nature, 537, 2016
|
|
7XIO
 
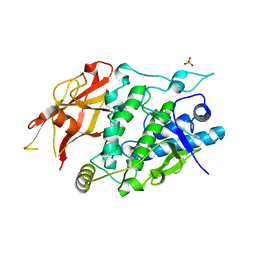 | | Crystal structure of TYR from Ralstonia | | Descriptor: | PHOSPHATE ION, Polyphenol oxidase | | Authors: | Sun, D.Y, Cui, P.P, Liao, L.J, Liu, X.K, Liu, B, Guo, Y, Feng, Z, Zhang, J, Li, X, Zeng, Z.X. | | Deposit date: | 2022-04-13 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of TYR from Ralstonia
To Be Published
|
|
5LQW
 
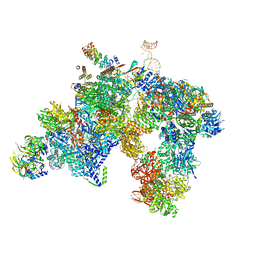 | | yeast activated spliceosome | | Descriptor: | Pre-mRNA leakage protein 1, Pre-mRNA-processing protein 45, Pre-mRNA-splicing factor 8, ... | | Authors: | Rauhut, R, Luehrmann, R. | | Deposit date: | 2016-08-17 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Molecular architecture of the Saccharomyces cerevisiae activated spliceosome
Science, 6306, 2016
|
|
5M3K
 
 | | A multi-component acyltransferase PhlABC from Pseudomonas protegens | | Descriptor: | 2,4-diacetylphloroglucinol biosynthesis protein PhlB, 2,4-diacetylphloroglucinol biosynthesis protein PhlC, PhlA, ... | | Authors: | Pavkov-Keller, T, Schmidt, N.G, Kroutil, W, Gruber, K. | | Deposit date: | 2016-10-15 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure and Catalytic Mechanism of a Bacterial Friedel-Crafts Acylase.
Chembiochem, 20, 2019
|
|
