6J0B
 
 | | Cryo-EM Structure of an Extracellular Contractile Injection System, PVC sheath-tube complex in extended state | | Descriptor: | Pvc1, Pvc2 | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
6J0C
 
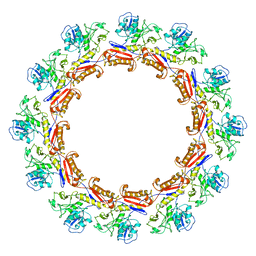 | | Cryo-EM Structure of an Extracellular Contractile Injection System, PVC sheath complex in contracted state | | Descriptor: | Pvc2 | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-23 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
6J0F
 
 | | Cryo-EM Structure of an Extracellular Contractile Injection System, PVC sheath/tube terminator in extended state | | Descriptor: | Pvc1, Pvc16 | | Authors: | Jiang, F, Li, N, Wang, X, Cheng, J, Huang, Y, Yang, Y, Yang, J, Cai, B, Wang, Y, Jin, Q, Gao, N. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM Structure and Assembly of an Extracellular Contractile Injection System.
Cell, 177, 2019
|
|
1UW0
 
 | |
8GOB
 
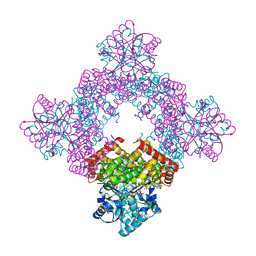 | | Crystal Structure of Glycerol Dehydrogenase in the presence of NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
8GOA
 
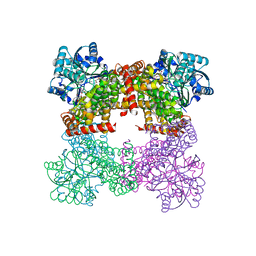 | | Crystal Structure of Glycerol Dehydrogenase in the absence of NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycerol dehydrogenase, ZINC ION | | Authors: | Park, T, Hoang, H.N, Kang, J.Y, Park, J, Mun, S.A, Jin, M, Yang, J, Jung, C.-H, Eom, S.H. | | Deposit date: | 2022-08-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insights into the flexible beta-hairpin of glycerol dehydrogenase.
Febs J., 290, 2023
|
|
6BA6
 
 | | Solution structure of Rap1b/talin complex | | Descriptor: | Ras-related protein Rap-1b, Talin-1 | | Authors: | Zhu, L, Yang, J, Qin, J. | | Deposit date: | 2017-10-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of Rap1b bound to talin reveals a pathway for triggering integrin activation.
Nat Commun, 8, 2017
|
|
4OK0
 
 | | Crystal structure of putative nucleotidyltransferase from H. pylori | | Descriptor: | Putative | | Authors: | Yoon, J.Y, Lee, S.J, Lee, B, Yang, J.K, Suh, S.W. | | Deposit date: | 2014-01-21 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of JHP933 from Helicobacter pylori J99 shows two-domain architecture with a DUF1814 family nucleotidyltransferase domain and a helical bundle domain.
Proteins, 82, 2014
|
|
5B6C
 
 | | Structural Details of Ufd1 binding to p97 | | Descriptor: | Peptide from Ubiquitin fusion degradation protein 1 homolog, Transitional endoplasmic reticulum ATPase | | Authors: | Le, L.T.M, Yang, J.K. | | Deposit date: | 2016-05-26 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Details of Ufd1 Binding to p97 and Their Functional Implications in ER-Associated Degradation
PLoS ONE, 11, 2016
|
|
3RBT
 
 | | Crystal structure of glutathione S-transferase Omega 3 from the silkworm Bombyx mori | | Descriptor: | GLYCEROL, Glutathione transferase o1 | | Authors: | Chen, B.-Y, Ma, X.-X, Tan, X, Yang, J.-P, Zhang, N.-N, Li, W.-F, Chen, Y, Xia, Q, Zhou, C.-Z. | | Deposit date: | 2011-03-29 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-guided activity restoration of the silkworm glutathione transferase Omega GSTO3-3
J.Mol.Biol., 412, 2011
|
|
3MWH
 
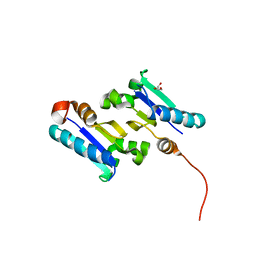 | | The 1.4 Ang crystal structure of the ArsD arsenic metallochaperone provides insights into its interactions with the ArsA ATPase | | Descriptor: | Arsenical resistance operon trans-acting repressor arsD, GLYCEROL | | Authors: | Ye, J, Ajees, A.A, Yang, J, Rosen, B.P. | | Deposit date: | 2010-05-05 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The 1.4 A crystal structure of the ArsD arsenic metallochaperone provides insights into its interaction with the ArsA ATPase.
Biochemistry, 49, 2010
|
|
6JE9
 
 | | Crystal structure of Nme1Cas9-sgRNA dimer mediated by double protein inhibitor AcrIIC3 monomers | | Descriptor: | AcrIIC3, CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-04 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDQ
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA | | Descriptor: | CRISPR-associated endonuclease Cas9, sgRNA | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JDV
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in catalytic state | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, MAGNESIUM ION, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JE3
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) with 5 nt overhang | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target DNA strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-03 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.931 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6JFU
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
5E84
 
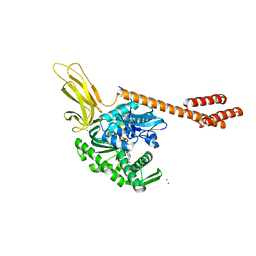 | | ATP-bound state of BiP | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, Q, Yang, J, Nune, M, Zong, Y, Zhou, L. | | Deposit date: | 2015-10-13 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Close and Allosteric Opening of the Polypeptide-Binding Site in a Human Hsp70 Chaperone BiP.
Structure, 23, 2015
|
|
8GQW
 
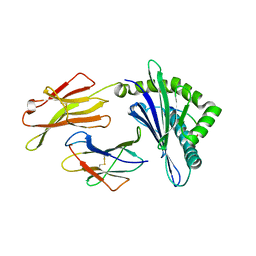 | | The Crystal Structures of a Swine SLA-2*HB01 Molecules Complexed with a CTL epitope from Asia1 serotype of Foot-and-mouth disease virus | | Descriptor: | Hu64, MHC class I antigen, beta 2 microglobulin | | Authors: | Feng, L, Gao, Y.Y, Sun, M.W, Li, Z.B, Zhang, Q, Yang, J, Qiao, C, Jin, H, Feng, H.S, Xian, Y.H, Qi, J.X, Gao, G.F, Liu, W.J, Gao, F.S. | | Deposit date: | 2022-08-31 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The Parallel Presentation of Two Functional CTL Epitopes Derived from the O and Asia 1 Serotypes of Foot-and-Mouth Disease Virus and Swine SLA-2*HB01: Implications for Universal Vaccine Development.
Cells, 11, 2022
|
|
8GQV
 
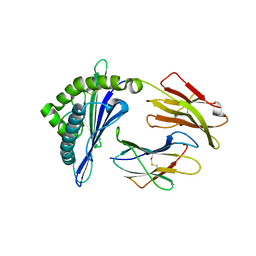 | | The Crystal Structures of a Swine SLA-2*HB01 Molecules Complexed with a CTL epitope from Asia1 serotype of Foot-and-mouth disease virus | | Descriptor: | As64, MHC class I antigen, beta 2 microglobulin | | Authors: | Feng, L, Gao, Y.Y, Sun, M.W, Li, Z.B, Zhang, Q, Yang, J, Qiao, C, Jin, H, Feng, H.S, Xian, Y.H, Qi, J.X, Gao, G.F, Liu, W.J, Gao, F.S. | | Deposit date: | 2022-08-31 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Parallel Presentation of Two Functional CTL Epitopes Derived from the O and Asia 1 Serotypes of Foot-and-Mouth Disease Virus and Swine SLA-2*HB01: Implications for Universal Vaccine Development.
Cells, 11, 2022
|
|
7CLD
 
 | | Crystal structure of T2R-TTL-Cevipabulin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[2,6-bis(fluoranyl)-4-[3-(methylamino)propoxy]phenyl]-5-chloranyl-N-[(2S)-1,1,1-tris(fluoranyl)propan-2-yl]-[1,2,4]triazolo[1,5-a]pyrimidin-7-amine, CALCIUM ION, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-07-20 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Cevipabulin-tubulin complex reveals a novel agent binding site on alpha-tubulin with tubulin degradation effect.
Sci Adv, 7, 2021
|
|
6KE1
 
 | | Crystal structure of TtCas1 | | Descriptor: | CRISPR-associated endonuclease Cas1 2 | | Authors: | Wang, Y.L, Li, J.Z, Yang, J, Wang, J.Y. | | Deposit date: | 2019-07-03 | | Release date: | 2019-12-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.39 Å) | | Cite: | Crystal structure of Cas1 in complex with branched DNA.
Sci China Life Sci, 63, 2020
|
|
6KC7
 
 | | Crystal structure of Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in seed-base paring state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*AP*AP*AP*GP*TP*T)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6KC8
 
 | | Crystal structure of WT Nme1Cas9 in complex with sgRNA and target DNA (ATATGATT PAM) in post-cleavage state | | Descriptor: | CRISPR-associated endonuclease Cas9, DNA (5'-D(*AP*TP*AP*TP*GP*AP*TP*TP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*AP*AP*TP*CP*AP*TP*AP*TP*GP*TP*A)-3'), ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-06-27 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
6KDV
 
 | | Crystal structure of TtCas1-DNA complex | | Descriptor: | CRISPR-associated endonuclease Cas1 2, DNA (5'-D(*GP*AP*GP*TP*CP*GP*AP*TP*GP*CP*TP*GP*GP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*CP*CP*AP*GP*CP*AP*TP*CP*GP*AP*CP*TP*C)-3') | | Authors: | Wang, Y.L, Li, J.Z, Yang, J, Wang, J.Y. | | Deposit date: | 2019-07-02 | | Release date: | 2019-12-18 | | Last modified: | 2020-05-06 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of Cas1 in complex with branched DNA.
Sci China Life Sci, 63, 2020
|
|
7DP8
 
 | | Crystal structure of T2R-TTL-Cevipabulin-eribulin complex | | Descriptor: | (1S,3S,6S,9S,12S,14R,16R,18S,20R,21R,22S,26R,29S,31R,32S,33R,35R,36S)-20-[(2S)-3-amino-2-hydroxypropyl]-21-methoxy-14-methyl-8,15-dimethylidene-2,19,30,34,37,39,40,41-octaoxanonacyclo[24.9.2.1~3,32~.1~3,33~.1~6,9~.1~12,16~.0~18,22~.0~29,36~.0~31,35~]hentetracontan-24-one (non-preferred name), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-[2,6-bis(fluoranyl)-4-[3-(methylamino)propoxy]phenyl]-5-chloranyl-N-[(2S)-1,1,1-tris(fluoranyl)propan-2-yl]-[1,2,4]triazolo[1,5-a]pyrimidin-7-amine, ... | | Authors: | Chen, L.J, Chen, Q, Yu, Y, Yang, J.H. | | Deposit date: | 2020-12-18 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | Cevipabulin-tubulin complex reveals a novel agent binding site on alpha-tubulin with tubulin degradation effect.
Sci Adv, 7, 2021
|
|
