7VUX
 
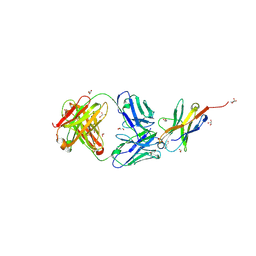 | | Complex structure of PD1 and 609A-Fab | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Huang, H, Zhu, Z, Zhao, J, Jiang, L, Yang, H, Deng, L, Meng, X, Ding, J, Yang, S, Zhao, L, Xu, W, Wang, X. | | Deposit date: | 2021-11-04 | | Release date: | 2021-11-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A strategy for the efficient construction of anti-PD1-based bispecific antibodies with desired IgG-like properties.
Mabs, 14, 2022
|
|
6J3O
 
 | | Crystal structure of the human PCAF bromodomain in complex with compound 12 | | Descriptor: | 3-methyl-2-[[(3~{R})-1-methylpiperidin-3-yl]amino]-5~{H}-pyrrolo[3,2-d]pyrimidin-4-one, Histone acetyltransferase KAT2B | | Authors: | Huang, L.Y, Li, H, Li, L.L, Niu, L, Seupel, R, Wu, C.Y, Li, G.B, Yu, Y.M, Brennan, P.E, Yang, S.Y. | | Deposit date: | 2019-01-05 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of Pyrrolo[3,2- d]pyrimidin-4-one Derivatives as a New Class of Potent and Cell-Active Inhibitors of P300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 62, 2019
|
|
7C7B
 
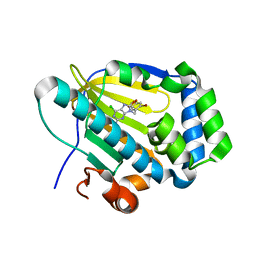 | | Crystal structure of human TRAP1 with SJT009 | | Descriptor: | 2-azanyl-9-[(6-bromanyl-1,3-benzodioxol-5-yl)methyl]-6-chloranyl-purin-8-ol, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Yang, S, Yoon, N.G, Park, E, Kim, S.Y, Kang, B.H, Lee, C, Kang, S. | | Deposit date: | 2020-05-24 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design and Synthesis of TRAP1 Selective Inhibitors: H-Bonding with Asn171 Residue in TRAP1 Increases Paralog Selectivity.
Acs Med.Chem.Lett., 12, 2021
|
|
7C7C
 
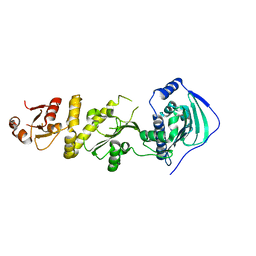 | | Crystal structure of human TRAP1 with SJT104 | | Descriptor: | 2-azanyl-9-[(4-bromanyl-2-fluoranyl-phenyl)methyl]-6-chloranyl-purin-8-ol, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Yang, S, Yoon, N.G, Park, E, Kim, S.Y, Kang, B.H, Lee, C, Kang, S. | | Deposit date: | 2020-05-24 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and Synthesis of TRAP1 Selective Inhibitors: H-Bonding with Asn171 Residue in TRAP1 Increases Paralog Selectivity.
Acs Med.Chem.Lett., 12, 2021
|
|
4EUF
 
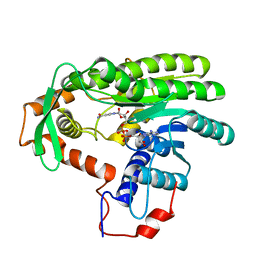 | | Crystal structure of Clostridium acetobutulicum trans-2-enoyl-CoA reductase in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative reductase CA_C0462, SODIUM ION | | Authors: | Hu, K, Zhao, M, Zhang, T, Yang, S, Ding, J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism
Biochem.J., 449, 2013
|
|
5WPS
 
 | | Crystal structure HpiC1 Y101F | | Descriptor: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.389 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6J3P
 
 | | Crystal structure of the human GCN5 bromodomain in complex with compound (R,R)-36n | | Descriptor: | 2-{[(3R,5R)-5-(2,3-dihydro-1,4-benzodioxin-6-yl)-1-methylpiperidin-3-yl]amino}-3-methyl-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, Histone acetyltransferase KAT2A | | Authors: | Huang, L.Y, Li, H, Niu, L, Wu, C.Y, Yu, Y.M, Li, L.L, Yang, S.Y. | | Deposit date: | 2019-01-05 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Discovery of Pyrrolo[3,2- d]pyrimidin-4-one Derivatives as a New Class of Potent and Cell-Active Inhibitors of P300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 62, 2019
|
|
5XDM
 
 | |
1L8J
 
 | | Crystal Structure of the Endothelial Protein C Receptor and Bound Phospholipid Molecule | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endothelial protein C receptor, ... | | Authors: | Oganesyan, V, Oganesyan, N, Terzyan, S, Dongfeng, Q, Dauter, Z, Esmon, N.L, Esmon, C.T. | | Deposit date: | 2002-03-20 | | Release date: | 2002-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the endothelial protein C receptor and a bound phospholipid.
J.Biol.Chem., 277, 2002
|
|
1JLK
 
 | | Crystal structure of the Mn(2+)-bound form of response regulator Rcp1 | | Descriptor: | MANGANESE (II) ION, Response regulator RCP1, SULFATE ION | | Authors: | Im, Y.J, Rho, S.-H, Park, C.-M, Yang, S.-S, Kang, J.-G, Lee, J.Y, Song, P.-S, Eom, S.H. | | Deposit date: | 2001-07-16 | | Release date: | 2002-03-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a cyanobacterial phytochrome response regulator.
Protein Sci., 11, 2002
|
|
7DMY
 
 | | The crystal structure of Cpd7 in complex with BPTF bromodomain | | Descriptor: | Nucleosome-remodeling factor subunit BPTF, tert-butyl 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-4-oxidanylidene-5,7-dihydropyrrolo[3,4-d]pyrimidine-6-carboxylate | | Authors: | Xiong, L, Guo, Y, Yang, S. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of selective BPTF bromodomain inhibitors by screening and structure-based optimization.
Biochem.Biophys.Res.Commun., 545, 2021
|
|
7DN4
 
 | | The crystal structure of Cpd8 in complex with BPTF bromodomain | | Descriptor: | 3-methyl-2-[[(3R,5R)-1-methyl-5-phenyl-piperidin-3-yl]amino]-6,7-dihydro-5H-cyclopenta[d]pyrimidin-4-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Xiong, L, Guo, Y, Yang, S. | | Deposit date: | 2020-12-08 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Discovery of selective BPTF bromodomain inhibitors by screening and structure-based optimization.
Biochem.Biophys.Res.Commun., 545, 2021
|
|
6IMC
 
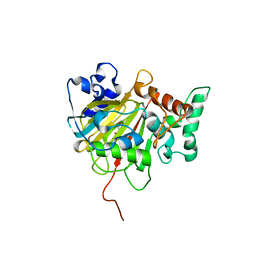 | | Crystal Structure of ALKBH1 in complex with Mn(II) and N-Oxalylglycine | | Descriptor: | MANGANESE (II) ION, N-OXALYLGLYCINE, Nucleic acid dioxygenase ALKBH1 | | Authors: | Zhang, M, Yang, S, Zhao, W, Li, H. | | Deposit date: | 2018-10-22 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Mammalian ALKBH1 serves as an N6-mA demethylase of unpairing DNA.
Cell Res., 30, 2020
|
|
8IXW
 
 | |
3LQU
 
 | |
7XGX
 
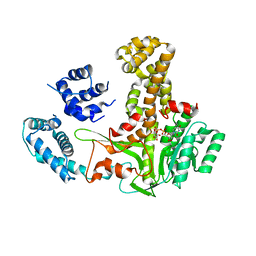 | | Glucosyltransferase | | Descriptor: | Lgt2, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Chen, T.T, Ouyang, S.Y. | | Deposit date: | 2022-04-07 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural Basis for the Action Mechanism of Legionella Glycosyltransferase.
Small Struct, 2023
|
|
7XGV
 
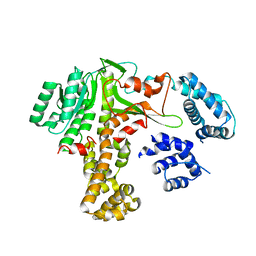 | | Legionella glucosyltransferase | | Descriptor: | Lgt2 | | Authors: | Chen, T.T, Ouyang, S.Y. | | Deposit date: | 2022-04-06 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Basis for the Action Mechanism of Legionella Glycosyltransferase.
Small Struct, 2023
|
|
5XEC
 
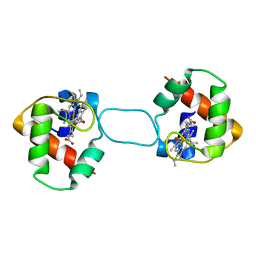 | | Heterodimer constructed from PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
6UZT
 
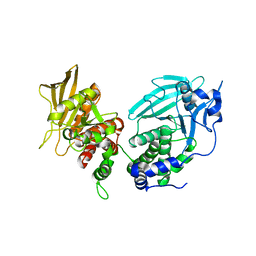 | | Crystal Structure of RPTP alpha | | Descriptor: | Receptor-type tyrosine-protein phosphatase alpha | | Authors: | Santelli, E, Wen, Y, Yang, S, Svensson, M.N.D, Stanford, S.M, Bottini, N. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | RPTP alpha phosphatase activity is allosterically regulated by the membrane-distal catalytic domain.
J.Biol.Chem., 295, 2020
|
|
7EQV
 
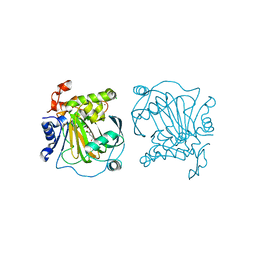 | | Crystal structure of JMJD2A complexed with 3,4-dihydroxybenzoic acid | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, CHLORIDE ION, Lysine-specific demethylase 4A, ... | | Authors: | Fang, W.-K, Yang, S.-M, Wang, W.-C. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Natural product myricetin is a pan-KDM4 inhibitor which with poly lactic-co-glycolic acid formulation effectively targets castration-resistant prostate cancer.
J.Biomed.Sci., 29, 2022
|
|
8H5S
 
 | | Crystal structure of Rv3400 from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Beta-phosphoglucomutase, CHLORIDE ION, ... | | Authors: | Singh, L, Karthikeyan, S, Thakur, K.G. | | Deposit date: | 2022-10-13 | | Release date: | 2023-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical and structural characterization reveals Rv3400 codes for beta-phosphoglucomutase in Mycobacterium tuberculosis.
Protein Sci., 33, 2024
|
|
7CAP
 
 | | Cyclic Lys48-linked triubiquitin | | Descriptor: | Ubiquitin, ZINC ION | | Authors: | Hiranyakorn, M, Yanaka, S, Satoh, T, Wilasri, T, Jityuti, B, Yagi-Utsumi, T, Kato, K. | | Deposit date: | 2020-06-09 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | NMR Characterization of Conformational Interconversions of Lys48-Linked Ubiquitin Chains.
Int J Mol Sci, 21, 2020
|
|
2JRV
 
 | | The third dimensional structure of mab198-bound pep.1 for autoimmune myasthenia gravis | | Descriptor: | PEPTIDE PEP.1 | | Authors: | Jung, H.H, Yi, H.J, Lee, S.K, Lee, J.Y, Jung, H.J, Yang, S.T, Eu, Y.-J, Im, S.-H, Kim, J.I. | | Deposit date: | 2007-06-29 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of immunotherapeutic peptides for autoimmune Myasthenia gravis
Biochemistry, 46, 2007
|
|
2JRW
 
 | | Solution structure of Cyclic extended Pep1(Cyc.ext.Pep.1) for autoimmune myasthenia gravis | | Descriptor: | Cyclic extended Pep.1 | | Authors: | Jung, H.H, Yi, H.J, Lee, S.K, Lee, J.Y, Jung, H.J, Yang, S.T, Eu, Y.-J, Im, S.-H, Kim, J.I. | | Deposit date: | 2007-06-29 | | Release date: | 2008-07-01 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of immunotherapeutic peptides for autoimmune Myasthenia gravis
Biochemistry, 46, 2007
|
|
1LQV
 
 | | Crystal structure of the Endothelial protein C receptor with phospholipid in the groove in complex with Gla domain of protein C. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endothelial protein C receptor, ... | | Authors: | Oganesyan, V, Oganesyan, N, Terzyan, S, Dongfeng, Q, Dauter, Z, Esmon, N.L, Esmon, C.T. | | Deposit date: | 2002-05-13 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the endothelial protein C receptor and a bound phospholipid.
J.Biol.Chem., 277, 2002
|
|
