1A8K
 
 | | CRYSTALLOGRAPHIC ANALYSIS OF HUMAN IMMUNODEFICIENCY VIRUS 1 PROTEASE WITH AN ANALOG OF THE CONSERVED CA-P2 SUBSTRATE: INTERACTIONS WITH FREQUENTLY OCCURRING GLUTAMIC ACID RESIDUE AT P2' POSITION OF SUBSTRATES | | Descriptor: | HIV PROTEASE, N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide | | Authors: | Weber, I.T, Wu, J, Adomat, J, Harrison, R.W, Kimmel, A.R, Wondrak, E.M, Louis, J.M. | | Deposit date: | 1998-03-27 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analysis of human immunodeficiency virus 1 protease with an analog of the conserved CA-p2 substrate -- interactions with frequently occurring glutamic acid residue at P2' position of substrates.
Eur.J.Biochem., 249, 1997
|
|
2HS2
 
 | | Crystal structure of M46L mutant of HIV-1 protease complexed with TMC114 (darunavir) | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weber, I.T, Kovalevsky, A.Y, Liu, F. | | Deposit date: | 2006-07-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Ultra-high Resolution Crystal Structure of HIV-1 Protease Mutant Reveals Two Binding Sites for Clinical Inhibitor TMC114.
J.Mol.Biol., 363, 2006
|
|
2HS1
 
 | | Ultra-high resolution X-ray crystal structure of HIV-1 protease V32I mutant with TMC114 (darunavir) inhibitor | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Weber, I.T, Kovalevsky, A.Y. | | Deposit date: | 2006-07-20 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Ultra-high Resolution Crystal Structure of HIV-1 Protease Mutant Reveals Two Binding Sites for Clinical Inhibitor TMC114.
J.Mol.Biol., 363, 2006
|
|
9B2H
 
 | | HIV-1 wild type protease with GRL-072-17A, a substituted tetrahydrofuran derivative based on Darunavir as P2 group | | Descriptor: | 2,5:6,9-dianhydro-1,3,7,8-tetradeoxy-4-O-({(2S,3R)-3-hydroxy-4-[(4-methoxybenzene-1-sulfonyl)(2-methylpropyl)amino]-1-phenylbutan-2-yl}carbamoyl)-L-gluco-nonitol, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2024-03-15 | | Release date: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Design of substituted tetrahydrofuran derivatives for HIV-1 protease inhibitors: synthesis, biological evaluation, and X-ray structural studies.
Org.Biomol.Chem., 2024
|
|
6PRF
 
 | | HIV-1 Protease multiple drug resistant clinical isolate mutant PR20 with GRL-14213A | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease, YTTRIUM ION | | Authors: | Kneller, D.W, Agniswamy, J, Weber, I.T. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor combats highly drug resistant mutant PR20.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
6B4N
 
 | | a hydroxymethyl functionality at the 4-position of the 2-phenyloxazole moiety of HIV-1 protease inhibitors involving the P2' ligands | | Descriptor: | CHLORIDE ION, Protease, SODIUM ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-09-27 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Design, Synthesis, Biological Evaluation, and X-ray Studies of HIV-1 Protease Inhibitors with Modified P2' Ligands of Darunavir.
ChemMedChem, 12, 2017
|
|
6VOD
 
 | | HIV-1 wild type protease with GRL-052-16A, a tricyclic cyclohexane fused tetrahydrofuranofuran (CHf-THF) derivative as the P2 ligand | | Descriptor: | (1R,3aS,5R,6S,7aR)-octahydro-1,6-epoxy-2-benzofuran-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2020-01-30 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure-Based Design of Highly Potent HIV-1 Protease Inhibitors Containing New Tricyclic Ring P2-Ligands: Design, Synthesis, Biological, and X-ray Structural Studies.
J.Med.Chem., 63, 2020
|
|
6VOE
 
 | | HIV-1 wild type protease with GRL-019-17A, a tricyclic cyclohexane fused tetrahydrofuranofuran (CHf-THF) derivative as the P2 ligand and a aminobenzothiazole(Abt)-based P2'-ligand | | Descriptor: | (1S,3aR,5S,6R,7aS)-octahydro-1,6-epoxy-2-benzofuran-5-yl {(2S,3R)-3-hydroxy-4-[(2-methylpropyl)({2-[(propan-2-yl)amino]-1,3-benzoxazol-6-yl}sulfonyl)amino]-1-phenylbutan-2-yl}carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2020-01-30 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-Based Design of Highly Potent HIV-1 Protease Inhibitors Containing New Tricyclic Ring P2-Ligands: Design, Synthesis, Biological, and X-ray Structural Studies.
J.Med.Chem., 63, 2020
|
|
8DCI
 
 | |
8DCH
 
 | | Crystal Structure of a highly resistant HIV-1 protease Clinical isolate PR10x with GRL-0519 (tris-tetrahydrofuran as P2 ligand) | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wong-Sam, A.E, Wang, Y.-F, Weber, I.T. | | Deposit date: | 2022-06-16 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | HIV-1 protease with 10 lopinavir and darunavir resistance mutations exhibits altered inhibition, structural rearrangements and extreme dynamics.
J.Mol.Graph.Model., 117, 2022
|
|
3BVB
 
 | | Cystal structure of HIV-1 Active Site Mutant D25N and inhibitor Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-01-05 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Effect of the Active Site D25N Mutation on the Structure, Stability, and Ligand Binding of the Mature HIV-1 Protease.
J.Biol.Chem., 283, 2008
|
|
3BVA
 
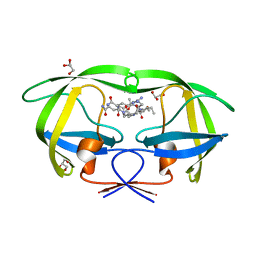 | | Cystal structure of HIV-1 Active Site Mutant D25N and p2-NC analog inhibitor | | Descriptor: | GLYCEROL, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, Protease (Retropepsin) | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-01-05 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of the Active Site D25N Mutation on the Structure, Stability, and Ligand Binding of the Mature HIV-1 Protease.
J.Biol.Chem., 283, 2008
|
|
5JFP
 
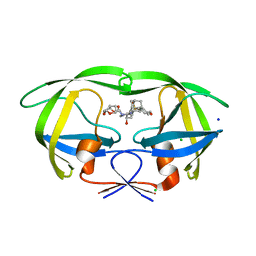 | | HIV-1 wild Type protease with GRL-097-13A (a Adamantane P1-Ligand with bis-THF in P2 and isobutylamine in P1') | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]butan-2-yl}carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.
J.Med.Chem., 59, 2016
|
|
7TO5
 
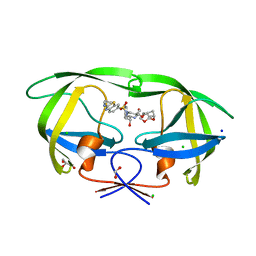 | | HIV-1 wild type protease with GRL-05816A, with C-4 substituted cyclohexane-fused bis-tetrahydrofuran (Chf-THF) derivatives as P2-ligand [diastereomer 1] | | Descriptor: | (1R,3aS,4S,6S,7aR)-octahydro-1,6-epoxy-2-benzofuran-4-yl [(2S,3R)-4-{[2-(cyclopropylamino)-1,3-benzothiazole-6-sulfonyl](2-methylpropyl)amino}-3-hydroxy-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Design, Synthesis and X-Ray Structural Studies of Potent HIV-1 Protease Inhibitors Containing C-4 Substituted Tricyclic Hexahydro-Furofuran Derivatives as P2 Ligands.
Chemmedchem, 17, 2022
|
|
7TO6
 
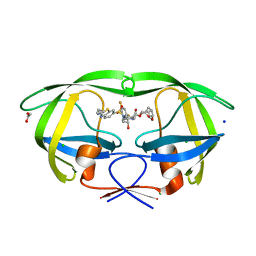 | | HIV-1 wild type protease with GRL-01717A, with C-4 substituted cyclohexane-fused bis-tetrahydrofuran (Chf-THF) derivatives as P2-ligand [diastereomer 2] | | Descriptor: | (1S,3aR,4R,6R,7aS)-octahydro-1,6-epoxy-2-benzofuran-4-yl [(2S,3R)-4-{[2-(cyclopropylamino)-1,3-benzothiazole-6-sulfonyl](2-methylpropyl)amino}-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Ghosh, A.K, Weber, I.T. | | Deposit date: | 2022-01-23 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Design, Synthesis and X-Ray Structural Studies of Potent HIV-1 Protease Inhibitors Containing C-4 Substituted Tricyclic Hexahydro-Furofuran Derivatives as P2 Ligands.
Chemmedchem, 17, 2022
|
|
7MYY
 
 | | Crystal Structure of HIV-1 PRS17 with GRL-142 | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Agniswamy, J, Weber, I.T. | | Deposit date: | 2021-05-22 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Novel HIV PR inhibitors with C4-substituted bis-THF and bis-fluoro-benzyl target the two active site mutations of highly drug resistant mutant PR S17 .
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7MYP
 
 | | Crystal Structure of HIV-1 PRS17 with GRL-44-10A | | Descriptor: | (3R,3aS,4R,6aR)-4-methoxyhexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Agniswamy, J, Weber, I.T. | | Deposit date: | 2021-05-21 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel HIV PR inhibitors with C4-substituted bis-THF and bis-fluoro-benzyl target the two active site mutations of highly drug resistant mutant PR S17 .
Biochem.Biophys.Res.Commun., 566, 2021
|
|
7N6T
 
 | |
7N6V
 
 | |
7N6X
 
 | |
5T2E
 
 | |
5T2Z
 
 | | Crystal Structure of Multi-drug Resistant HIV-1 Protease PR-S17 in Complex with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Agniswamy, J, Weber, I.T. | | Deposit date: | 2016-08-24 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Studies of a Rationally Selected Multi-Drug Resistant HIV-1 Protease Reveal Synergistic Effect of Distal Mutations on Flap Dynamics.
PLoS ONE, 11, 2016
|
|
1AGX
 
 | | REFINED CRYSTAL STRUCTURE OF ACINETOBACTER GLUTAMINASIFICANS GLUTAMINASE-ASPARAGINASE | | Descriptor: | GLUTAMINASE-ASPARAGINASE | | Authors: | Lubkowski, J, Wlodawer, A, Housset, D, Weber, I.T, Ammon, H.L, Murphy, K.C, Swain, A.L. | | Deposit date: | 1994-07-13 | | Release date: | 1994-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Refined crystal structure of Acinetobacter glutaminasificans glutaminase-asparaginase.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
5UOV
 
 | | HIV-1 wild Type protease with GRL-1118A , an isophthalamide-derived P2-P3 ligand with the sulfonamide isostere as the P2' group | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-02-01 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Design of novel HIV-1 protease inhibitors incorporating isophthalamide-derived P2-P3 ligands: Synthesis, biological evaluation and X-ray structural studies of inhibitor-HIV-1 protease complex.
Bioorg. Med. Chem., 25, 2017
|
|
3LJP
 
 | | Crystal structure of choline oxidase V464A mutant | | Descriptor: | Choline oxidase, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Finnegan, S, Agniswamy, J, Weber, I.T, Giovanni, G. | | Deposit date: | 2010-01-26 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of valine 464 in the flavin oxidation reaction catalyzed by choline oxidase.
Biochemistry, 49, 2010
|
|
