4MYU
 
 | |
4MRX
 
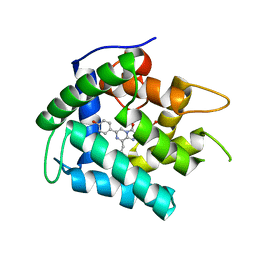 | | Crystal Structure of Y138F obelin mutant from Obelia longissima at 1.72 Angstrom resolution | | Descriptor: | C2-HYDROPEROXY-COELENTERAZINE, Obelin | | Authors: | Natashin, P.V, Ding, W, Eremeeva, E.V, Markova, S.V, Lee, J, Vysotski, E.S, Liu, Z.J. | | Deposit date: | 2013-09-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.718 Å) | | Cite: | Structures of the Ca2+-regulated photoprotein obelin Y138F mutant before and after bioluminescence support the catalytic function of a water molecule in the reaction.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4N39
 
 | |
4EIH
 
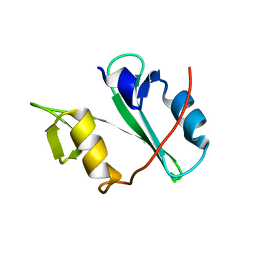 | | Crystal structure of Arg SH2 domain | | Descriptor: | Abelson tyrosine-protein kinase 2, CHLORIDE ION | | Authors: | Liu, W, MacGrath, S.M, Koleske, A.J, Boggon, T.J. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Two Amino Acid Residues Confer Different Binding Affinities of Abelson Family Kinase Src Homology 2 Domains for Phosphorylated Cortactin.
J.Biol.Chem., 289, 2014
|
|
4EIN
 
 | | Crystal structure of mouse thymidylate synthase in binary complex with a substrate analogue and strong inhibitor, N(4)-hydroxy-2'-deoxycytidine-5'-monophosphate | | Descriptor: | 2'-deoxy-N-hydroxycytidine 5'-(dihydrogen phosphate), GLYCEROL, Thymidylate synthase | | Authors: | Dowiercial, A, Jarmula, A, Rypniewski, W.R, Wilk, P, Kierdaszuk, B, Rode, W. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of complexes of mouse thymidylate synthase
crystallized with N4-OH-dCMP alone or in the presence of
N5,10-methylenetetrahydrofolate
PTERIDINES, 2013
|
|
4CCH
 
 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in an open binary complex with d5SICS as templating nucleotide | | Descriptor: | 5'-D(*AP*AP*CP*LHOP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP* C)-3', 5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*DOC)-3', DNA POLYMERASE I, ... | | Authors: | Betz, K, Malyshev, D.A, Lavergne, T, Welte, W, Diederichs, K, Romesberg, F.E, Marx, A. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights Into DNA Replication without Hydrogen Bonds.
J.Am.Chem.Soc., 135, 2013
|
|
4CE7
 
 | | Crystal structure of a novel unsaturated beta-glucuronyl hydrolase enzyme, belonging to family GH105, involved in ulvan degradation | | Descriptor: | NITRATE ION, UNSATURATED 3S-RHAMNOGLYCURONYL HYDROLASE | | Authors: | Nyvall Collen, P, Jeudy, A, Groisillier, A, Coutinho, P.M, Helbert, W, Czjzek, M. | | Deposit date: | 2013-11-09 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Unsaturated Beta-Glucuronyl Hydrolase Involved in Ulvan Degradation Unveils the Versatility of Stereochemistry Requirements in Family Gh105.
J.Biol.Chem., 289, 2014
|
|
4BT1
 
 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR | | Authors: | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | Deposit date: | 2013-06-12 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (16 Å) | | Cite: | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BQH
 
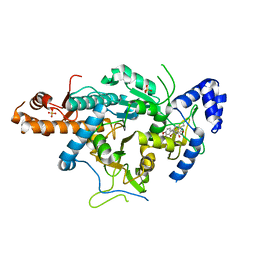 | | Crystal structure of the uridine diphosphate N-acetylglucosamine pyrophosphorylase from Trypanosoma brucei in complex with inhibitor | | Descriptor: | (3S)-3-[2-(1,3-benzodioxol-5-yl)-2-oxidanylidene-ethyl]-4-bromanyl-5-methyl-3-oxidanyl-1H-indol-2-one, SULFATE ION, UDP-N-ACETYLGLUCOSAMINE PYROPHOSPHORYLASE | | Authors: | Fang, W, Raimi, O.G, vanAalten, D.M.F. | | Deposit date: | 2013-05-30 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Novel Allosteric Inhibitor of the Uridine Diphosphate N-Acetylglucosamine Pyrophosphorylase from Trypanosoma Brucei.
Acs Chem.Biol., 8, 2013
|
|
4BV5
 
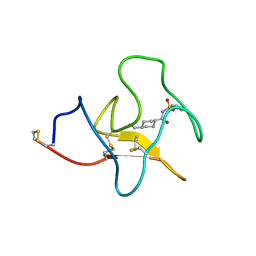 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 4-(aminomethyl)-N-(benzenesulfonyl)cyclohexanecarboxamide, APOLIPOPROTEIN(A) | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4N14
 
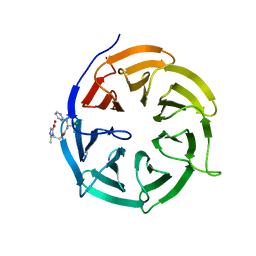 | | Crystal structure of Cdc20 and apcin complex | | Descriptor: | 2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethyl [(1R)-2,2,2-trichloro-1-(pyrimidin-2-ylamino)ethyl]carbamate, Cell division cycle protein 20 homolog | | Authors: | Luo, X, Tian, W, Yu, H. | | Deposit date: | 2013-10-03 | | Release date: | 2014-08-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C.
Nature, 514, 2014
|
|
4BT0
 
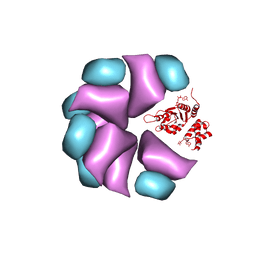 | | MuB is an AAAplus ATPase that forms helical filaments to control target selection for DNA transposition | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, TRANSCRIPTIONAL REGULATOR | | Authors: | Mizuno, N, Dramicanin, M, Mizuuchi, M, Adam, J, Wang, Y, Han, Y.W, Yang, W, Steven, A.C, Mizuuchi, K, Ramon-Maiques, S. | | Deposit date: | 2013-06-12 | | Release date: | 2013-07-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Mub is an Aaa+ ATPase that Forms Helical Filaments to Control Target Selection for DNA Transposition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BXL
 
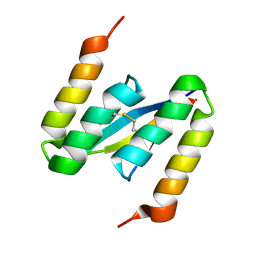 | | Structure of alpha-synuclein in complex with an engineered binding protein | | Descriptor: | ALPHA SYNUCLEIN, AS69 | | Authors: | Mirecka, E.A, Shaykhalishahi, H, Lecher, J, Stoldt, M, Hoyer, W. | | Deposit date: | 2013-07-12 | | Release date: | 2014-05-21 | | Method: | SOLUTION NMR | | Cite: | Sequestration of a Beta-Hairpin for Control of Alpha-Synuclein Aggregation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4BOL
 
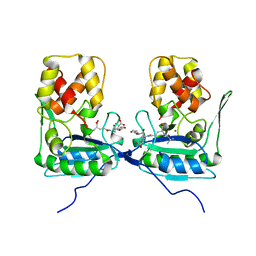 | | Crystal structure of AmpDh2 from Pseudomonas aeruginosa in complex with pentapeptide | | Descriptor: | AMPDH2, D-alanyl-N-[(2S,6R)-6-amino-6-carboxy-1-{[(1R)-1-carboxyethyl]amino}-1-oxohexan-2-yl]-D-glutamine, ZINC ION | | Authors: | Artola-Recolons, C, Martinez-Caballero, S, Lee, M, Carrasco-Lopez, C, Hesek, D, Spink, E.E, Lastochkin, E, Zhang, W, Hellman, L.M, Boggess, B, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-05-21 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
4LJL
 
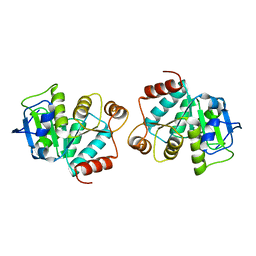 | |
4BML
 
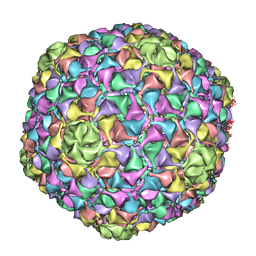 | | C-alpha backbone trace of major capsid protein gp39 found in marine virus Syn5. | | Descriptor: | MAJOR CAPSID PROTEIN | | Authors: | Gipson, P, Baker, M.L, Raytcheva, D, Haase-Pettingell, C, Piret, J, King, J, Chiu, W. | | Deposit date: | 2013-05-09 | | Release date: | 2014-05-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Protruding Knob-Like Proteins Violate Local Symmetries in an Icosahedral Marine Virus.
Nat.Commun., 5, 2014
|
|
4BSS
 
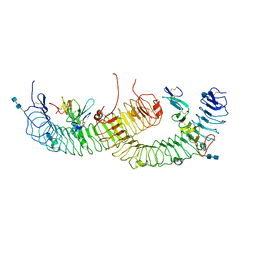 | | Structure of the ectodomain of LGR5 in complex with R-spondin-1 (Fu1Fu2) in P21 crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LEUCINE-RICH REPEAT-CONTAINING G-PROTEIN COUPLED RECEPTOR 5, ... | | Authors: | Peng, W.C, de Lau, W, Forneris, F, Granneman, J.C.M, Huch, M, Clevers, H, Gros, P. | | Deposit date: | 2013-06-11 | | Release date: | 2013-06-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of Stem Cell Growth Factor R-Spondin 1 in Complex with the Ectodomain of its Receptor Lgr5.
Cell Rep., 3, 2013
|
|
4LJK
 
 | |
4BQJ
 
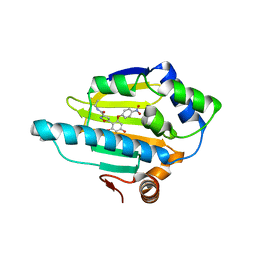 | | structure of HSP90 with an inhibitor bound | | Descriptor: | 5-[2,4-dihydroxy-6-(4-nitrophenoxy)phenyl]-N-ethyl-1,2-oxazole-3-carboxamide, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Casale, E, Brasca, M.G, Mantegani, S, Amboldi, N, Bindi, S, Caronni, D, Ceccarelli, W, Colombo, N, DePonti, A, Donati, D, Ermoli, A, Fachin, G, Felder, E.R, Ferguson, R.D, Fiorelli, C, Guanci, M, Isacchi, A, Pesenti, E, Polucci, P, Riceputi, L, Sola, F, Visco, C, Zuccotto, F, Fogliatto, G. | | Deposit date: | 2013-05-30 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Nms-E973 as Novel, Selective and Potent Inhibitor of Heat Shock Protein 90 (Hsp90).
Bioorg.Med.Chem., 21, 2013
|
|
4LMQ
 
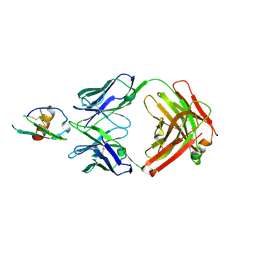 | | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12 | | Descriptor: | Stromal cell-derived factor 1, hu30D8 Fab heavy chain, hu30D8 Fab light chain | | Authors: | Zhong, Z, Wang, J, Li, B, Xiang, H, Ultsch, M, Coons, M, Wong, T, Chiang, N.Y, Clark, S, Clark, R, Quintana, L, Gribling, P, Suto, E, Barck, K, Corpuz, R, Yao, J, Takkar, R, Lee, W.P, Damico-Beyer, L.A, Carano, R.D, Adams, C, Kelley, R.F, Wang, W, Ferrara, N. | | Deposit date: | 2013-07-10 | | Release date: | 2013-08-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.773 Å) | | Cite: | Development and Preclinical Characterization of a Humanized Antibody Targeting CXCL12.
Clin.Cancer Res., 19, 2013
|
|
4MG5
 
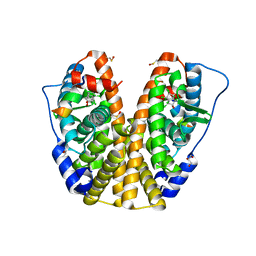 | | Crystal structure of hERa-LBD (Y537S) in complex with chlordecone | | Descriptor: | Estrogen receptor, GLYCEROL, Nuclear receptor coactivator 1, ... | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural and functional profiling of environmental ligands for estrogen receptors.
Environ.Health Perspect., 122, 2014
|
|
4CA9
 
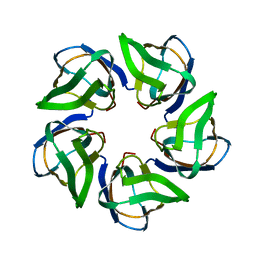 | | Structure of the Nucleoplasmin-like N-terminal domain of Drosophila FKBP39 | | Descriptor: | 39 KDA FK506-BINDING NUCLEAR PROTEIN | | Authors: | Artero, J, Forsyth, T, Callow, P, Watson, A.A, Zhang, W, Laue, E.D, Edlich-Muth, C, Przewloka, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-10-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Pentameric Nucleoplasmin Fold is Present in Drosophila Fkbp39 and a Large Number of Chromatin-Related Proteins.
J.Mol.Biol., 427, 2015
|
|
4LAK
 
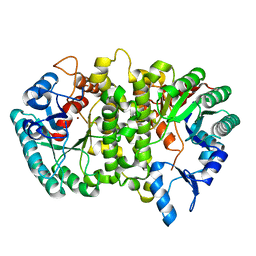 | | Crystal structure of Cordyceps militaris IDCase D323N mutant in apo form | | Descriptor: | Uracil-5-carboxylate decarboxylase, ZINC ION | | Authors: | Xu, S, Li, W, Zhu, J, Wang, R, Li, Z, Xu, G.L, Ding, J. | | Deposit date: | 2013-06-20 | | Release date: | 2013-10-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structures of isoorotate decarboxylases reveal a novel catalytic mechanism of 5-carboxyl-uracil decarboxylation and shed light on the search for DNA decarboxylase.
Cell Res., 23, 2013
|
|
4BVD
 
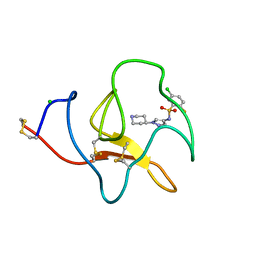 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 5-chloro-2-fluoro-N-[1-(4-piperidyl)pyrazol-4-yl]benzenesulfonamide, APOLIPOPROTEIN(A), CHLORIDE ION | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
4BV7
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | Descriptor: | 3-(4-piperidyl)propanoic acid, ACETATE ION, APOLIPOPROTEIN(A) | | Authors: | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | Deposit date: | 2013-06-25 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
