7TOJ
 
 | | Crystal structure of carbohydrate esterase CspAcXE, apoenzyme | | Descriptor: | CHLORIDE ION, SGNH/GDSL hydrolase family protein | | Authors: | Stogios, P.J, Skarina, T, Di Leo, R, Jurak, E, Master, E. | | Deposit date: | 2022-01-24 | | Release date: | 2022-04-13 | | Last modified: | 2022-11-02 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Elucidating Sequence and Structural Determinants of Carbohydrate Esterases for Complete Deacetylation of Substituted Xylans.
Molecules, 27, 2022
|
|
6MN5
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(3)-IVa, H154A mutant, in complex with gentamicin C1A | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Kim, Y, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
6MGJ
 
 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, apoenzyme | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Nocek, B, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
6MN1
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with gentamicin-CoA | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aminoglycoside N(3)-acetyltransferase, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with gentamicin-CoA
To Be Published
|
|
3V75
 
 | | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680 | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Stogios, P.J, Xu, X, Cui, H, Kudritska, M, Tan, K, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-12-20 | | Release date: | 2012-05-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of putative orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis ma-4680
To be Published
|
|
6M8U
 
 | | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, prenylated-FMN complex | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Flavin prenyltransferase UbiX, PHOSPHATE ION | | Authors: | Stogios, P.J, Skarina, T, Khusnutidinova, A, Wawrzak, Z, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2018-08-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, prenylated-FMN complex
To Be Published
|
|
6M8V
 
 | | Crystal structure of UbiX-like FMN prenyltransferase MJ0101 from Methanocaldococcus jannaschii, FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Khusnutdinova, A, Wawrzak, Z, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2018-08-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Crystal structure of UbiX-like FMN prenyltransferase MJ0101 from Methanocaldococcus jannaschii, FMN complex
To Be Published
|
|
6AOJ
 
 | | Crystal structure of Legionella pneumophila effector Ceg4 with N-terminal yeast Hog1p sequence | | Descriptor: | CHLORIDE ION, Ceg4, MAGNESIUM ION | | Authors: | Stogios, P.J, Nocek, B, Cuff, M.E, Evdokimova, E, Egorova, O, Yim, V, Di Leo, R, Savchenko, A. | | Deposit date: | 2017-08-16 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | TheLegionella pneumophilaeffector Ceg4 is a phosphotyrosine phosphatase that attenuates activation of eukaryotic MAPK pathways.
J. Biol. Chem., 293, 2018
|
|
6M8T
 
 | | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, FMN complex | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin prenyltransferase UbiX, PHOSPHATE ION | | Authors: | Stogios, P.J, Skarina, T, Khusnutidnova, A, Wawrzak, Z, Yakunin, A.F, Savchenko, A. | | Deposit date: | 2018-08-22 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of UbiX-like FMN prenyltransferase AF1214 from Archaeoglobus fulgidus, FMN complex
To Be Published
|
|
6AOK
 
 | | Crystal structure of Legionella pneumophila effector Ceg4 with N-terminal TEV protease cleavage sequence | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Ceg4, ... | | Authors: | Stogios, P.J, Cuff, M.E, Nocek, B, Evdokimova, E, Egorova, O, Yim, V, Di Leo, R, Savchenko, A. | | Deposit date: | 2017-08-16 | | Release date: | 2018-01-10 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | TheLegionella pneumophilaeffector Ceg4 is a phosphotyrosine phosphatase that attenuates activation of eukaryotic MAPK pathways.
J. Biol. Chem., 293, 2018
|
|
6MMZ
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H29A mutant apoenzyme | | Descriptor: | Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, SULFATE ION | | Authors: | Stogios, P.J, Skarina, T, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
6MGK
 
 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, in complex with XLX xyloglucan | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Nocek, B, Arnal, G, Brumer, H, Savchenko, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
6BNC
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, Thr315Ala mutant di-zinc and PEG complex | | Descriptor: | CHLORIDE ION, POLYETHYLENE GLYCOL (N=34), Phosphoethanolamine transferase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Substrate Recognition by a Colistin Resistance Enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 13, 2018
|
|
6MN0
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, H168A mutant in complex with acetyl-CoA | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETYL COENZYME *A, Aminoglycoside N(3)-acetyltransferase, ... | | Authors: | Stogios, P.J, Skarina, T, Zu, X, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and molecular rationale for the diversification of resistance mediated by the Antibiotic_NAT family.
Commun Biol, 5, 2022
|
|
6C8Q
 
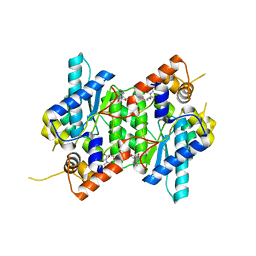 | | Crystal structure of NAD synthetase (NadE) from Enterococcus faecalis in complex with NAD+ | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Stogios, P.J, Skarina, T, McChesney, C, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-25 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.583 Å) | | Cite: | To be published
To Be Published
|
|
6MSW
 
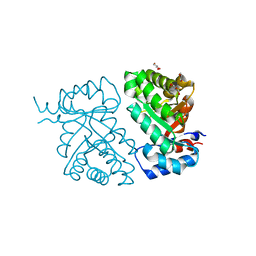 | | Crystal structure of BH1352 2-deoxyribose-5-phosphate from Bacillus halodurans, K184L mutant | | Descriptor: | Deoxyribose-phosphate aldolase, GLYCEROL | | Authors: | Stogios, P.J, Skarina, T, Kim, T, Yim, V, Yakunin, A, Savchenko, A. | | Deposit date: | 2018-10-18 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.169 Å) | | Cite: | Rational engineering of 2-deoxyribose-5-phosphate aldolases for the biosynthesis of (R)-1,3-butanediol.
J.Biol.Chem., 295, 2020
|
|
4FEV
 
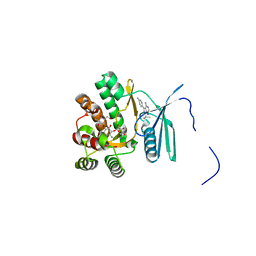 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor pyrazolopyrimidine PP1 | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
6MGL
 
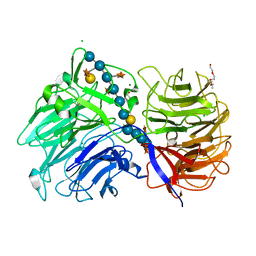 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, D60A mutant in complex with XXLG and XGXXLG xyloglucan | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Skarina, T, Arnal, G, Watanabe, N, Brumer, H, Savchenko, A. | | Deposit date: | 2018-09-14 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
6MIJ
 
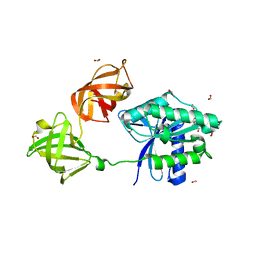 | | Crystal structure of EF-Tu from Acinetobacter baumannii in complex with Mg2+ and GDP | | Descriptor: | Elongation factor Tu, FORMIC ACID, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-19 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | To be published
To Be Published
|
|
6MN2
 
 | | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with sisomicin-CoA | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, Aminoglycoside N(3)-acetyltransferase, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Michalska, K, Xu, Z, Yim, V, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Crystal structure of meta-AAC0038, an environmental aminoglycoside resistance enzyme, mutant H168A in abortive complex with sisomicin-CoA
To Be Published
|
|
4GKH
 
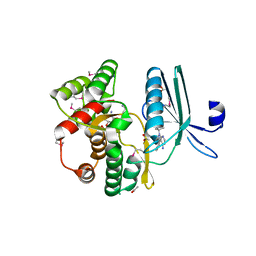 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NA-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4GKI
 
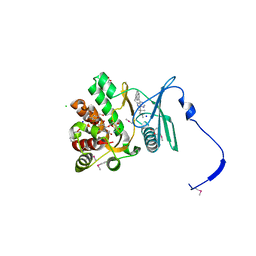 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NM-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4FEW
 
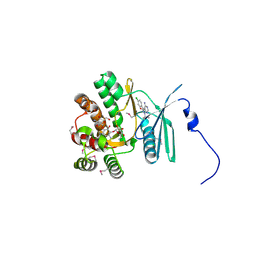 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor pyrazolopyrimidine PP2 | | Descriptor: | 1-TERT-BUTYL-3-(4-CHLORO-PHENYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
6BND
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, Thr315Ala mutant mono-zinc and phosphoethanolamine complex | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, POLYETHYLENE GLYCOL (N=34), Phosphoethanolamine transferase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Substrate Recognition by a Colistin Resistance Enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 13, 2018
|
|
6BNE
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, phosphate-bound complex | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Substrate recognition by a colistin resistance enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 2018
|
|
