1UPU
 
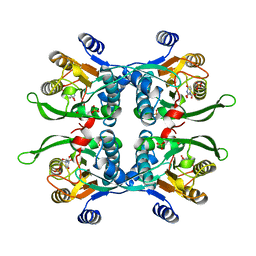 | | STRUCTURE OF THE URACIL PHOSPHORIBOSYLTRANSFERASE, MUTANT C128V, BOUND TO PRODUCT URIDINE-1-MONOPHOSPHATE (UMP) | | Descriptor: | PHOSPHATE ION, URACIL PHOSPHORIBOSYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE | | Authors: | Schumacher, M.A, Carter, D, Scott, D, Roos, D, Ullman, B, Brennan, R.G. | | Deposit date: | 1998-04-16 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Toxoplasma gondii uracil phosphoribosyltransferase reveal the atomic basis of pyrimidine discrimination and prodrug binding.
EMBO J., 17, 1998
|
|
5J4L
 
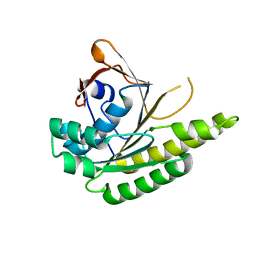 | | Apo-structure of humanised RadA-mutant humRadA22F | | Descriptor: | CHLORIDE ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
8BBZ
 
 | |
8BHU
 
 | | Crystal Structure of SilF (Ag(I) form | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SILVER ION, SULFATE ION, ... | | Authors: | Lithgo, R.M, Carr, S.B, Quigley, A.M, Scott, D.J. | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of SilF (apo form)
To Be Published
|
|
5KDD
 
 | | Apo-structure of humanised RadA-mutant humRadA22 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
1UIU
 
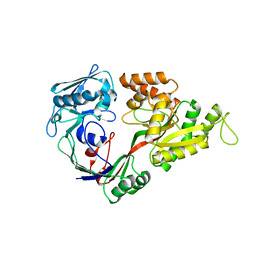 | | Crystal structures of the liganded and unliganded nickel binding protein NikA from Escherichia coli (Nickel unliganded form) | | Descriptor: | Nickel-binding periplasmic protein | | Authors: | Heddle, J, Scott, D.J, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2003-07-22 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the liganded and unliganded nickel-binding protein NikA from Escherichia coli
J.Biol.Chem., 278, 2003
|
|
1UIV
 
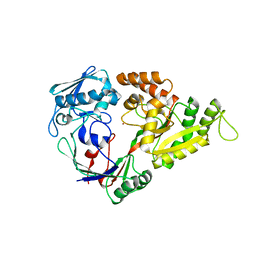 | | Crystal structures of the liganded and unliganded nickel binding protein NikA from Escherichia coli (Nickel liganded form) | | Descriptor: | NICKEL (II) ION, Nickel-binding periplasmic protein | | Authors: | Heddle, J, Scott, D.J, Unzai, S, Park, S.-Y, Tame, J.R.H. | | Deposit date: | 2003-07-22 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of the liganded and unliganded nickel-binding protein NikA from Escherichia coli
J.Biol.Chem., 278, 2003
|
|
6XTW
 
 | | HumRadA33F in complex with peptidic inhibitor 6 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION, ~{N}-[2-[(2~{S})-2-[[(1~{S})-1-(4-methoxyphenyl)ethyl]carbamoyl]pyrrolidin-1-yl]-2-oxidanylidene-ethyl]quinoline-2-carboxamide | | Authors: | Fischer, G, Marsh, M.E, Scott, D.E, Coyne, A.G, Skidmore, J, Abell, C, Hyvonen, M. | | Deposit date: | 2020-01-16 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A small-molecule inhibitor of the BRCA2-RAD51 interaction modulates RAD51 assembly and potentiates DNA damage-induced cell death.
Cell Chem Biol, 28, 2021
|
|
5V83
 
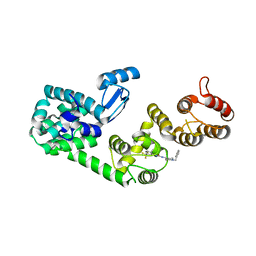 | | Structure of DCN1 bound to NAcM-HIT | | Descriptor: | Lysozyme,DCN1-like protein 1 chimera, N-(1-benzylpiperidin-4-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
7B6W
 
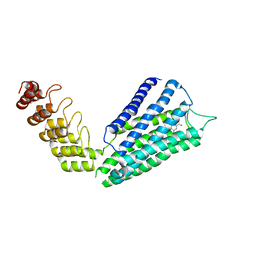 | | Crystal structure of the human alpha1B adrenergic receptor in complex with inverse agonist (+)-cyclazosin | | Descriptor: | Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor,Alpha-1B adrenergic receptor,alpha1B adrenergic receptor, [(4~{a}~{R},8~{a}~{S})-4-(4-azanyl-6,7-dimethoxy-quinazolin-2-yl)-2,3,4~{a},5,6,7,8,8~{a}-octahydroquinoxalin-1-yl]-(furan-2-yl)methanone | | Authors: | Deluigi, M, Morstein, L, Hilge, M, Schuster, M, Merklinger, L, Klipp, A, Scott, D.J, Plueckthun, A. | | Deposit date: | 2020-12-08 | | Release date: | 2022-01-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.873 Å) | | Cite: | Crystal structure of the alpha 1B -adrenergic receptor reveals molecular determinants of selective ligand recognition.
Nat Commun, 13, 2022
|
|
5V89
 
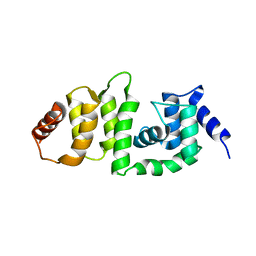 | | Structure of DCN4 PONY domain bound to CUL1 WHB | | Descriptor: | Cullin-1, DCN1-like protein 4 | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
5V86
 
 | | Structure of DCN1 bound to NAcM-OPT | | Descriptor: | Lysozyme,DCN1-like protein 1, N-benzyl-N-(1-butylpiperidin-4-yl)-N'-(3,4-dichlorophenyl)urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
5V88
 
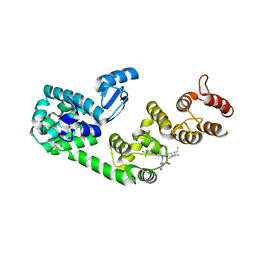 | | Structure of DCN1 bound to NAcM-COV | | Descriptor: | Lysozyme,DCN1-like protein 1, N-{2-[({1-[(2R)-pentan-2-yl]piperidin-4-yl}{[3-(trifluoromethyl)phenyl]carbamoyl}amino)methyl]phenyl}propanamide | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
8BWV
 
 | |
3FN1
 
 | | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. | | Descriptor: | NEDD8-activating enzyme E1 catalytic subunit, NEDD8-conjugating enzyme UBE2F | | Authors: | Huang, D.T, Ayrault, O, Hunt, H.W, Taherbhoy, A.M, Duda, D.M, Scott, D.C, Borg, L.A, Neale, G, Murray, P.J, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2008-12-22 | | Release date: | 2009-03-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification
Mol.Cell, 33, 2009
|
|
6BG3
 
 | | Structure of (3S,4S)-1-benzyl-4-(3-(3-(trifluoromethyl)phenyl)ureido)piperidin-3-yl acetate bound to DCN1 | | Descriptor: | Endolysin, DCN1-like protein 1 chimera, N-{(3S,4S)-1-benzyl-3-[(1S)-1-hydroxyethoxy]piperidin-4-yl}-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-10-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
1DTO
 
 | | CRYSTAL STRUCTURE OF THE COMPLETE TRANSACTIVATION DOMAIN OF E2 PROTEIN FROM THE HUMAN PAPILLOMAVIRUS TYPE 16 | | Descriptor: | REGULATORY PROTEIN E2 | | Authors: | Antson, A.A, Burns, J.E, Moroz, O.V, Scott, D.J, Sanders, C.M, Bronstein, I.B, Dodson, G.G, Wilson, K.S, Maitland, N. | | Deposit date: | 2000-01-13 | | Release date: | 2000-02-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the intact transactivation domain of the human papillomavirus E2 protein.
Nature, 403, 2000
|
|
3IOB
 
 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 1.80 Ang resolution in complex with 5'-deoxy-5'-thioadenosine | | Descriptor: | 5'-thioadenosine, ETHANOL, GLYCEROL, ... | | Authors: | Ciulli, A, Scott, D.E, Abell, C. | | Deposit date: | 2009-08-14 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Fragment-Based Approach to Probing Adenosine Recognition Sites by Using Dynamic Combinatorial Chemistry
Chembiochem, 10, 2009
|
|
3DQV
 
 | | Structural Insights into NEDD8 Activation of Cullin-RING Ligases: Conformational Control of Conjugation | | Descriptor: | Cullin-5, NEDD8, Rbx1, ... | | Authors: | Duda, D.M, Borg, L.A, Scott, D.C, Hunt, H.W, Hammel, M, Schulman, B.A. | | Deposit date: | 2008-07-09 | | Release date: | 2008-09-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation.
Cell(Cambridge,Mass.), 134, 2008
|
|
5KYK
 
 | | Covalent GTP-competitive inhibitors of KRAS G12C: Guanosine bisphosphonate Analogs | | Descriptor: | 5'-O-[(R)-[({2-[(chloroacetyl)amino]ethyl}sulfamoyl)methyl](hydroxy)phosphoryl]guanosine, GTPase KRas | | Authors: | Xiong, Y, Lu, J, Hunter, J, Li, L, Scott, D, Manandhar, A, Gondi, S, Westover, K.D, Gray, N.S. | | Deposit date: | 2016-07-21 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.702 Å) | | Cite: | Covalent Guanosine Mimetic Inhibitors of G12C KRAS.
ACS Med Chem Lett, 8, 2017
|
|
3IOD
 
 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 1.75 Ang resolution in complex with 5'-deoxy-5'-((3-nitrobenzyl)disulfanyl)-adenosine | | Descriptor: | (2R,3R,4S,5S)-2-(6-amino-9H-purin-9-yl)-5-{[(3-nitrobenzyl)disulfanyl]methyl}tetrahydrofuran-3,4-diol, ETHANOL, GLYCEROL, ... | | Authors: | Ciulli, A, Scott, D.E, Abell, C. | | Deposit date: | 2009-08-14 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Fragment-Based Approach to Probing Adenosine Recognition Sites by Using Dynamic Combinatorial Chemistry
Chembiochem, 10, 2009
|
|
3IOE
 
 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 1.95 Ang resolution in complex with 5'-deoxy-5'-((R)-3,4-dihydroxybutylthio)-adenosine | | Descriptor: | 5'-S-[(3R)-3,4-dihydroxybutyl]-5'-thioadenosine, ETHANOL, GLYCEROL, ... | | Authors: | Ciulli, A, Scott, D.E, Abell, C. | | Deposit date: | 2009-08-14 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Fragment-Based Approach to Probing Adenosine Recognition Sites by Using Dynamic Combinatorial Chemistry
Chembiochem, 10, 2009
|
|
1Y2O
 
 | | Structure of N-terminal domain IRSp53/BAIAP2 | | Descriptor: | BAI1-associated protein 2 isoform 1 | | Authors: | Millard, T.H, Bompard, G, Heung, M.-Y, Dafforn, T.R, Scott, D.J, Machesky, L.M, Futterer, K. | | Deposit date: | 2004-11-23 | | Release date: | 2005-02-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of filopodia formation induced by the IRSp53/MIM homology domain of human IRSp53
Embo J., 24, 2005
|
|
5JZK
 
 | | The Structure of Ultra Stable Green Fluorescent Protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yong, K.J, Gunn, N.J, Scott, D.J, Griffin, M.D.W. | | Deposit date: | 2016-05-17 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Ultra-Stable, Monomeric Green Fluorescent Protein For Direct Volumetric Imaging of Whole Organs Using CLARITY.
Sci Rep, 8, 2018
|
|
3IOC
 
 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 2.50 Ang resolution in complex with 5'-deoxy-5'-(benzyldisulfanyl)-adenosine | | Descriptor: | (2R,3R,4S,5S)-2-(6-amino-9H-purin-9-yl)-5-[(benzyldisulfanyl)methyl]tetrahydrofuran-3,4-diol, GLYCEROL, Pantothenate synthetase | | Authors: | Ciulli, A, Scott, D.E, Abell, C. | | Deposit date: | 2009-08-14 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Fragment-Based Approach to Probing Adenosine Recognition Sites by Using Dynamic Combinatorial Chemistry
Chembiochem, 10, 2009
|
|
