1DC0
 
 | |
2O60
 
 | |
3GOF
 
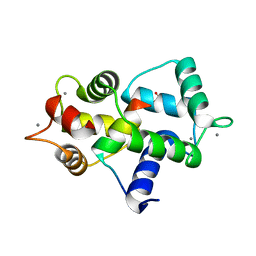 | | Calmodulin bound to peptide from macrophage nitric oxide synthase | | Descriptor: | CALCIUM ION, Calmodulin, Nitric oxide synthase, ... | | Authors: | Ng, H.L, Greenstein, A, Marletta, M, Wand, A.J, Alber, T. | | Deposit date: | 2009-03-19 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural diversity in calmodulin recognition of nitric oxide synthases
To be Published
|
|
3GP2
 
 | |
4OJ0
 
 | | mCardinal V218E | | Descriptor: | Fluorescent protein FP480 | | Authors: | Ataie, N, Ng, H. | | Deposit date: | 2014-01-20 | | Release date: | 2014-03-19 | | Last modified: | 2014-05-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein.
Nat.Methods, 11, 2014
|
|
1PJG
 
 | | RNA/DNA Hybrid Decamer of CAAAGAAAAG/CTTTTCTTTG | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', CALCIUM ION | | Authors: | Kopka, M.L, Lavelle, L, Han, G.W, Ng, H.-L, Dickerson, R.E. | | Deposit date: | 2003-06-02 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | An unusual sugar conformation in the structure of an RNA/DNA decamer of the polypurine tract may affect recognition by RNase H.
J.Mol.Biol., 334, 2003
|
|
1PJO
 
 | | Crystal Structure of an RNA/DNA hybrid of HIV-1 PPT | | Descriptor: | 5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3', 5'-R(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3', MAGNESIUM ION | | Authors: | Kopka, M.L, Lavelle, L, Han, G.W, Ng, H.-L, Dickerson, R.E. | | Deposit date: | 2003-06-03 | | Release date: | 2003-12-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | An Unusual Sugar Conformation in the Structure of an RNA/DNA
Decamer of the Polypurine Tract May Affect Recognition by RNase H
J.Mol.Biol., 334, 2003
|
|
2MHF
 
 | |
2N7G
 
 | |
4WUU
 
 | | Structure of ESK1 in complex with HLA-A*0201/WT1 | | Descriptor: | ARG-MET-PHE-PRO-ASN-ALA-PRO-TYR-LEU, Beta-2-microglobulin, ESK1, ... | | Authors: | Ataie, N.J, Ng, H.L. | | Deposit date: | 2014-11-03 | | Release date: | 2015-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.047 Å) | | Cite: | Structure of a TCR-Mimic Antibody with Target Predicts Pharmacogenetics.
J.Mol.Biol., 428, 2016
|
|
3OXH
 
 | | Mycobacterium tuberculosis kinase inhibitor homolog RV0577 | | Descriptor: | CHLORIDE ION, PARA-MERCURY-BENZENESULFONIC ACID, RV0577 PROTEIN, ... | | Authors: | Echols, N, Flynn, E.M, Stephenson, S, Ng, H.-L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-21 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biophysical Characterization of the Mycobacterium tuberculosis Protein Rv0577, a Protein Associated with Neutral Red Staining of Virulent Tuberculosis Strains and Homologue of the Streptomyces coelicolor Protein KbpA.
Biochemistry, 2017
|
|
5KDD
 
 | | Apo-structure of humanised RadA-mutant humRadA22 | | Descriptor: | DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JEC
 
 | | Apo-structure of humanised RadA-mutant humRadA33F | | Descriptor: | CHLORIDE ION, DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JFG
 
 | | Structure of humanised RadA-mutant humRadA22F in complex with peptide FHTA | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, DNA repair and recombination protein RadA, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-19 | | Release date: | 2016-10-26 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
3ORI
 
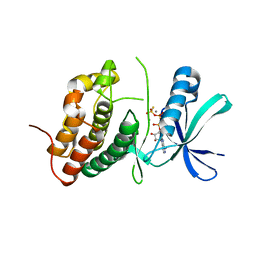 | | Mycobacterium tuberculosis PknB kinase domain L33D mutant (crystal form 1) | | Descriptor: | MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine protein kinase | | Authors: | Lombana, T.N, Echols, N, Good, M.C, Thomsen, N.D, Ng, H.-L, Alber, T, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2010-09-07 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric activation mechanism of the Mycobacterium tuberculosis receptor Ser/Thr protein kinase, PknB.
Structure, 18, 2010
|
|
6WNN
 
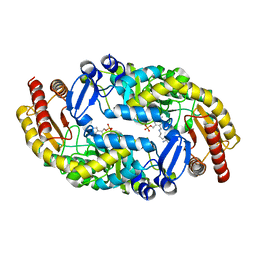 | | Bacillus subtilis BioA in complex with amino donor L-Lys | | Descriptor: | (2S)-2-amino-6-[[3-hydroxy-2-methyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]hexanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase | | Authors: | Souza, S.A, Ng, H.L. | | Deposit date: | 2020-04-22 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Substrate divergence in 7,8-diaminopelargonic acid synthesis: mutagenesis and computational studies of L-lysine dependent Bacillus subtilis BioA
To Be Published
|
|
7LUG
 
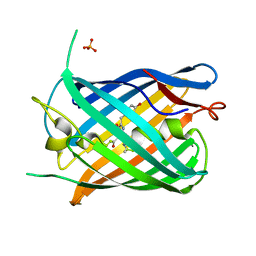 | | Crystal structure of the pnRFP B30Y mutant | | Descriptor: | PHOSPHATE ION, Red Fluorescent pnRFP B30Y mutant | | Authors: | Huang, M, Ng, H.L, Pang, Y, Zhang, S, Fan, Y, Yeh, H, Xiong, Y, Li, X, Ai, H. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development, Characterization, and Structural Analysis of a Genetically Encoded Red Fluorescent Peroxynitrite Biosensor
To Be Published
|
|
7LQO
 
 | | Crystal structure of a genetically encoded red fluorescent peroxynitrite biosensor, pnRFP | | Descriptor: | PHOSPHATE ION, red fluorescent peroxynitrite biosensor pnRFP | | Authors: | Huang, M, Ng, H.L, Pang, Y, Zhang, S, Fan, Y, Yeh, H, Xiong, Y, Li, X, Ai, H. | | Deposit date: | 2021-02-14 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Development, Characterization, and Structural Analysis of a Genetically Encoded Red Fluorescent Peroxynitrite Biosensor
To Be Published
|
|
5L8V
 
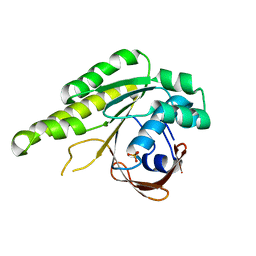 | | Apo-structure of humanised RadA-mutant humRadA4 | | Descriptor: | DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-08 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LB4
 
 | | Apo-structure of humanised RadA-mutant humRadA14 | | Descriptor: | DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LBI
 
 | | Apo-structure of humanised RadA-mutant humRadA3 | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-16 | | Release date: | 2016-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5LB2
 
 | | Apo-structure of humanised RadA-mutant humRadA2 | | Descriptor: | DNA repair and recombination protein RadA | | Authors: | Marsh, M, Fischer, G, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-06-15 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5J4K
 
 | | Structure of humanised RadA-mutant humRadA22F in complex with 1-Indane-6-carboxylic acid | | Descriptor: | 2,3-dihydro-1H-indene-2-carboxylic acid, CALCIUM ION, DNA repair and recombination protein RadA, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-26 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.346 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5J4H
 
 | | Structure of humanised RadA-mutant humRadA22F in complex with indole-6-carboxylic acid | | Descriptor: | 1H-indole-6-carboxylic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-19 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JED
 
 | | Apo-structure of humanised RadA-mutant humRadA28 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
